| Reaction Details |
|---|
 | Report a problem with these data |
| Target | [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 4, mitochondrial |
|---|
| Ligand | BDBM50472831 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_152460 (CHEMBL761449) |
|---|
| IC50 | 25±n/a nM |
|---|
| Citation |  Aicher, TD; Anderson, RC; Gao, J; Shetty, SS; Coppola, GM; Stanton, JL; Knorr, DC; Sperbeck, DM; Brand, LJ; Vinluan, CC; Kaplan, EL; Dragland, CJ; Tomaselli, HC; Islam, A; Lozito, RJ; Liu, X; Maniara, WM; Fillers, WS; DelGrande, D; Walter, RE; Mann, WR Secondary amides of (R)-3,3,3-trifluoro-2-hydroxy-2-methylpropionic acid as inhibitors of pyruvate dehydrogenase kinase. J Med Chem43:236-49 (2000) [PubMed] Article Aicher, TD; Anderson, RC; Gao, J; Shetty, SS; Coppola, GM; Stanton, JL; Knorr, DC; Sperbeck, DM; Brand, LJ; Vinluan, CC; Kaplan, EL; Dragland, CJ; Tomaselli, HC; Islam, A; Lozito, RJ; Liu, X; Maniara, WM; Fillers, WS; DelGrande, D; Walter, RE; Mann, WR Secondary amides of (R)-3,3,3-trifluoro-2-hydroxy-2-methylpropionic acid as inhibitors of pyruvate dehydrogenase kinase. J Med Chem43:236-49 (2000) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 4, mitochondrial |
|---|
| Name: | [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 4, mitochondrial |
|---|
| Synonyms: | 2.7.11.2 | PDK4_RAT | Pdk4 | Pyruvate dehydrogenase kinase isoform 4 | [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 4, mitochondrial |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 46681.07 |
|---|
| Organism: | Rattus norvegicus |
|---|
| Description: | ChEMBL_104818 |
|---|
| Residue: | 412 |
|---|
| Sequence: | MKAARFVMRSASSLGNAGLVPREVELFSRYSPSPLSMKQLLDFGSENACERTSFSFLRQE
LPVRLANILKEIDILPEHLVNTPSVQLVKSWYIQSLMDLVEFHEKSPEDQKVLSDFVDTL
VKVRNRHHNVVPTMAQGILEYKDNCTVDPVTNQNLQYFLDRFYMNRISTRMLMNQHILIF
SDSKTGNPSHIGSIDPNCDVVAVVEDAFECAKMLCDQYYLTSPELKLTQVNGKFPGQPIH
IVYVPSHLHHMLFELFKNAMRATVEHQENRPFLTPVEATVVLGKEDLTIKISDRGGGVPL
RITDRLFSYTYSTAPTPVMDNSRNAPLAGFGYGLPISRLYAKYFQGDLNLYSMSGYGTDA
IIYLKALSSESIEKLPVFNKSAFKHYQMSSEADDWCIPSKEPKNLSKEKLAV
|
|
|
|---|
| BDBM50472831 |
|---|
| n/a |
|---|
| Name | BDBM50472831 |
|---|
| Synonyms: | CHEMBL84271 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H21F3N2O4S |
|---|
| Mol. Mass. | 430.441 |
|---|
| SMILES | C[C@@H]1CN(CCN1S(=O)(=O)c1cccc2ccccc12)C(=O)[C@@](C)(O)C(F)(F)F |
|---|
| Structure | 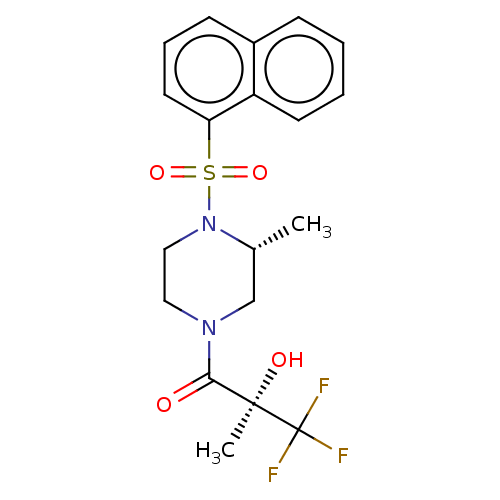
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Aicher, TD; Anderson, RC; Gao, J; Shetty, SS; Coppola, GM; Stanton, JL; Knorr, DC; Sperbeck, DM; Brand, LJ; Vinluan, CC; Kaplan, EL; Dragland, CJ; Tomaselli, HC; Islam, A; Lozito, RJ; Liu, X; Maniara, WM; Fillers, WS; DelGrande, D; Walter, RE; Mann, WR Secondary amides of (R)-3,3,3-trifluoro-2-hydroxy-2-methylpropionic acid as inhibitors of pyruvate dehydrogenase kinase. J Med Chem43:236-49 (2000) [PubMed] Article
Aicher, TD; Anderson, RC; Gao, J; Shetty, SS; Coppola, GM; Stanton, JL; Knorr, DC; Sperbeck, DM; Brand, LJ; Vinluan, CC; Kaplan, EL; Dragland, CJ; Tomaselli, HC; Islam, A; Lozito, RJ; Liu, X; Maniara, WM; Fillers, WS; DelGrande, D; Walter, RE; Mann, WR Secondary amides of (R)-3,3,3-trifluoro-2-hydroxy-2-methylpropionic acid as inhibitors of pyruvate dehydrogenase kinase. J Med Chem43:236-49 (2000) [PubMed] Article