| Reaction Details |
|---|
 | Report a problem with these data |
| Target | HIV-1 protease |
|---|
| Ligand | BDBM12897 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_517306 (CHEMBL1031359) |
|---|
| Ki | 0.072000±n/a nM |
|---|
| Citation |  Jorissen, RN; Reddy, GS; Ali, A; Altman, MD; Chellappan, S; Anjum, SG; Tidor, B; Schiffer, CA; Rana, TM; Gilson, MK Additivity in the analysis and design of HIV protease inhibitors. J Med Chem52:737-54 (2009) [PubMed] Article Jorissen, RN; Reddy, GS; Ali, A; Altman, MD; Chellappan, S; Anjum, SG; Tidor, B; Schiffer, CA; Rana, TM; Gilson, MK Additivity in the analysis and design of HIV protease inhibitors. J Med Chem52:737-54 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| HIV-1 protease |
|---|
| Name: | HIV-1 protease |
|---|
| Synonyms: | HIV-1 | HIV-1 protease | protease |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 10795.19 |
|---|
| Organism: | Human immunodeficiency virus |
|---|
| Description: | ChEMBL_118439 |
|---|
| Residue: | 99 |
|---|
| Sequence: | PQVTLWQRPLVTIKIGGQLKEALLDTGADDTVLEEMSLPGRWKPKMIGGIGGFIKVRQYD
QILIEICGHKAIGTVLIGPTPVNIIGRNLLTQIGCTLNF
|
|
|
|---|
| BDBM12897 |
|---|
| n/a |
|---|
| Name | BDBM12897 |
|---|
| Synonyms: | (5S)-N-[(1S,2R)-1-Benzyl-3-[[(3-fluoro-4-methoxyphenyl)-sulfonyl](isobutyl)amino]-2-hydroxypropyl]-2-oxo-3-[(3-trifluoro-methyl)phenyl)]oxazolidine-5-carboxamide | (5S)-N-[(2S,3R)-4-[(3-fluoro-4-methoxybenzene)(2-methylpropyl)sulfonamido]-3-hydroxy-1-phenylbutan-2-yl]-2-oxo-3-[3-(trifluoromethyl)phenyl]-1,3-oxazolidine-5-carboxamide | N-Aryl-oxazolidinone-5-carboxamide Analogue 27d |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C32H35F4N3O7S |
|---|
| Mol. Mass. | 681.695 |
|---|
| SMILES | COc1ccc(cc1F)S(=O)(=O)N(CC(C)C)C[C@@H](O)[C@H](Cc1ccccc1)NC(=O)[C@@H]1CN(C(=O)O1)c1cccc(c1)C(F)(F)F |r| |
|---|
| Structure | 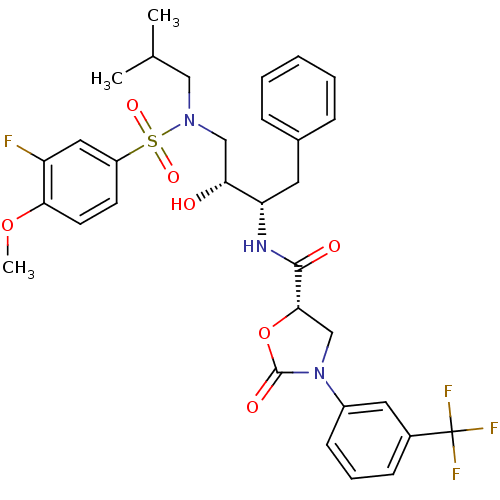
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Jorissen, RN; Reddy, GS; Ali, A; Altman, MD; Chellappan, S; Anjum, SG; Tidor, B; Schiffer, CA; Rana, TM; Gilson, MK Additivity in the analysis and design of HIV protease inhibitors. J Med Chem52:737-54 (2009) [PubMed] Article
Jorissen, RN; Reddy, GS; Ali, A; Altman, MD; Chellappan, S; Anjum, SG; Tidor, B; Schiffer, CA; Rana, TM; Gilson, MK Additivity in the analysis and design of HIV protease inhibitors. J Med Chem52:737-54 (2009) [PubMed] Article