| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Muscarinic acetylcholine receptor M1/M2/M3/M4/M5 |
|---|
| Ligand | BDBM50001884 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_138211 (CHEMBL748440) |
|---|
| Ki | 25±n/a nM |
|---|
| Citation |  Liégeois, JF; Bruhwyler, J; Damas, J; Nguyen, TP; Chleide, EM; Mercier, MG; Rogister, FA; Delarge, JE New pyridobenzodiazepine derivatives as potential antipsychotics: synthesis and neurochemical study. J Med Chem36:2107-14 (1993) [PubMed] Liégeois, JF; Bruhwyler, J; Damas, J; Nguyen, TP; Chleide, EM; Mercier, MG; Rogister, FA; Delarge, JE New pyridobenzodiazepine derivatives as potential antipsychotics: synthesis and neurochemical study. J Med Chem36:2107-14 (1993) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Muscarinic acetylcholine receptor M1/M2/M3/M4/M5 |
|---|
| Name: | Muscarinic acetylcholine receptor M1/M2/M3/M4/M5 |
|---|
| Synonyms: | Muscarinic acetylcholine receptor |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of ChEMBL is 1821274 |
|---|
| Components: | This complex has 5 components. |
|---|
| Component 1 |
| Name: | Muscarinic acetylcholine receptor M1 |
|---|
| Synonyms: | ACM1_RAT | Cholinergic, muscarinic M1 | Chrm-1 | Chrm1 | cholinergic receptor, muscarinic 1 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 51390.46 |
|---|
| Organism: | RAT |
|---|
| Description: | P08482 |
|---|
| Residue: | 460 |
|---|
| Sequence: | MNTSVPPAVSPNITVLAPGKGPWQVAFIGITTGLLSLATVTGNLLVLISFKVNTELKTVN
NYFLLSLACADLIIGTFSMNLYTTYLLMGHWALGTLACDLWLALDYVASNASVMNLLLIS
FDRYFSVTRPLSYRAKRTPRRAALMIGLAWLVSFVLWAPAILFWQYLVGERTVLAGQCYI
QFLSQPIITFGTAMAAFYLPVTVMCTLYWRIYRETENRARELAALQGSETPGKGGGSSSS
SERSQPGAEGSPESPPGRCCRCCRAPRLLQAYSWKEEEEEDEGSMESLTSSEGEEPGSEV
VIKMPMVDSEAQAPTKQPPKSSPNTVKRPTKKGRDRGGKGQKPRGKEQLAKRKTFSLVKE
KKAARTLSAILLAFILTWTPYNIMVLVSTFCKDCVPETLWELGYWLCYVNSTVNPMCYAL
CNKAFRDTFRLLLLCRWDKRRWRKIPKRPGSVHRTPSRQC
|
|
|
|---|
| Component 2 |
| Name: | Muscarinic acetylcholine receptor M3 |
|---|
| Synonyms: | ACM3_RAT | Cholinergic, muscarinic M3 | Chrm-3 | Chrm3 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 66086.66 |
|---|
| Organism: | RAT |
|---|
| Description: | Cholinergic, muscarinic M3 CHRM3 RAT::P08483 |
|---|
| Residue: | 589 |
|---|
| Sequence: | MTLHSNSTTSPLFPNISSSWVHSPSEAGLPLGTVTQLGSYNISQETGNFSSNDTSSDPLG
GHTIWQVVFIAFLTGFLALVTIIGNILVIVAFKVNKQLKTVNNYFLLSLACADLIIGVIS
MNLFTTYIIMNRWALGNLACDLWLSIDYVASNASVMNLLVISFDRYFSITRPLTYRAKRT
TKRAGVMIGLAWVISFVLWAPAILFWQYFVGKRTVPPGECFIQFLSEPTITFGTAIAAFY
MPVTIMTILYWRIYKETEKRTKELAGLQASGTEAEAENFVHPTGSSRSCSSYELQQQGVK
RSSRRKYGRCHFWFTTKSWKPSAEQMDQDHSSSDSWNNNDAAASLENSASSDEEDIGSET
RAIYSIVLKLPGHSSILNSTKLPSSDNLQVSNEDLGTVDVERNAHKLQAQKSMGDGDNCQ
KDFTKLPIQLESAVDTGKTSDTNSSADKTTATLPLSFKEATLAKRFALKTRSQITKRKRM
SLIKEKKAAQTLSAILLAFIITWTPYNIMVLVNTFCDSCIPKTYWNLGYWLCYINSTVNP
VCYALCNKTFRTTFKTLLLCQCDKRKRRKQQYQQRQSVIFHKRVPEQAL
|
|
|
|---|
| Component 3 |
| Name: | Muscarinic acetylcholine receptor M4 |
|---|
| Synonyms: | ACM4_RAT | Cholinergic, muscarinic M4 | Chrm-4 | Chrm4 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 52841.70 |
|---|
| Organism: | RAT |
|---|
| Description: | Cholinergic, muscarinic M4 CHRM4 RAT::P08485 |
|---|
| Residue: | 477 |
|---|
| Sequence: | MNFTPVNGSSANQSVRLVTAAHNHLETVEMVFIATVTGSLSLVTVVGNILVMLSIKVNRQ
LQTVNNYFLFSLGCADLIIGAFSMNLYTLYIIKGYWPLGAVVCDLWLALDYVVSNASVMN
LLIISFDRYFCVTKPLTYPARRTTKMAGLMIAAAWVLSFVLWAPAILFWQFVVGKRTVPD
NQCFIQFLSNPAVTFGTAIAAFYLPVVIMTVLYIHISLASRSRVHKHRPEGPKEKKAKTL
AFLKSPLMKPSIKKPPPGGASREELRNGKLEEAPPPALPPPPRPVPDKDTSNESSSGSAT
QNTKERPPTELSTAEATTPALPAPTLQPRTLNPASKWSKIQIVTKQTGNECVTAIEIVPA
TPAGMRPAANVARKFASIARNQVRKKRQMAARERKVTRTIFAILLAFILTWTPYNVMVLV
NTFCQSCIPERVWSIGYWLCYVNSTINPACYALCNATFKKTFRHLLLCQYRNIGTAR
|
|
|
|---|
| Component 4 |
| Name: | Muscarinic acetylcholine receptor M5 |
|---|
| Synonyms: | ACM5_RAT | Cholinergic, muscarinic M5 | Chrm-5 | Chrm5 | Muscarinic acetylcholine receptor | Muscarinic acetylcholine receptor M5 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 60161.80 |
|---|
| Organism: | RAT |
|---|
| Description: | Cholinergic, muscarinic M5 CHRM5 RAT::P08911 |
|---|
| Residue: | 531 |
|---|
| Sequence: | MEGESYNESTVNGTPVNHQALERHGLWEVITIAVVTAVVSLMTIVGNVLVMISFKVNSQL
KTVNNYYLLSLACADLIIGIFSMNLYTTYILMGRWVLGSLACDLWLALDYVASNASVMNL
LVISFDRYFSITRPLTYRAKRTPKRAGIMIGLAWLVSFILWAPAILCWQYLVGKRTVPPD
ECQIQFLSEPTITFGTAIAAFYIPVSVMTILYCRIYRETEKRTKDLADLQGSDSVAEAKK
REPAQRTLLRSFFSCPRPSLAQRERNQASWSSSRRSTSTTGKTTQATDLSADWEKAEQVT
TCSSYPSSEDEAKPTTDPVFQMVYKSEAKESPGKESNTQETKETVVNTRTENSDYDTPKY
FLSPAAAHRLKSQKCVAYKFRLVVKADGTQETNNGCRKVKIMPCSFPVSKDPSTKGPDPN
LSHQMTKRKRMVLVKERKAAQTLSAILLAFIITWTPYNIMVLVSTFCDKCVPVTLWHLGY
WLCYVNSTINPICYALCNRTFRKTFKLLLLCRWKKKKVEEKLYWQGNSKLP
|
|
|
|---|
| Component 5 |
| Name: | Muscarinic acetylcholine receptor M2 |
|---|
| Synonyms: | ACM2_RAT | Cholinergic, muscarinic M2 | Chrm-2 | Chrm2 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 51555.53 |
|---|
| Organism: | RAT |
|---|
| Description: | P10980 |
|---|
| Residue: | 466 |
|---|
| Sequence: | MNNSTNSSNNGLAITSPYKTFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNY
FLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFD
RYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVEDGECYIQF
FSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRIKKEKKEPVANQDPVSPSLVQGRI
VKPNNNNMPGGDGGLEHNKIQNGKAPRDGVTENCVQGEEKESSNDSTSVSAVASNMRDDE
ITQDENTVSTSLGHSRDDNSKQTCIKIVTKAQKGDVCTPTSTTVELVGSSGQNGDEKQNI
VARKIVKMTKQPAKKKPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNT
VWTIGYWLCYINSTINPACYALCNATFKKTFKHLLMCHYKNIGATR
|
|
|
|---|
| BDBM50001884 |
|---|
| n/a |
|---|
| Name | BDBM50001884 |
|---|
| Synonyms: | 2-[4-(4-Methyl-benzyl)-piperazin-1-yl]-1-(2-methyl-2,3-dihydro-indol-1-yl)-ethanone | 3-Hydroxy-2-phenyl-propionic acid 8-methyl-8-aza-bicyclo[3.2.1]oct-3-yl ester | 3-chloro-6-(4-methyl-piperazin-1-yl)-5H-dibenzo[b,e][1,4]diazepine | 8-Chloro-11-(4-methyl-piperazin-1-yl)-5H-dibenzo[b,e][1,4]diazepine (Clozapine) | 8-Chloro-11-(4-methyl-piperazin-1-yl)-5H-dibenzo[b,e][1,4]diazepine( Clozepine ) | 8-Chloro-11-(4-methyl-piperazin-1-yl)-5H-dibenzo[b,e][1,4]diazepine(Ciozapine) | 8-Chloro-11-(4-methyl-piperazin-1-yl)-5H-dibenzo[b,e][1,4]diazepine(Clopazine) | 8-chloro-11-(4-methyl-piperazin-1-yl)-10H-dibenzo[b,e][1,4]diazepine | 8-chloro-11-(4-methylpiperazin-1-yl)-5H-dibenzo[b,e][1,4]diazepine | CHEMBL42 | CHEMBL538973 | CLOZAPINE | CLOZAPINE, 8-CHLORO-11-(4-METHYL-PIPERAZIN-1-YL)-5H-DIBENZO[B,E][1,4]DIAZEPINE | CLOZARIL | HF 1854 | US10167256, Clozapine | US10752588, Compound Clozapine | US11498896, Compound Clozapine |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H19ClN4 |
|---|
| Mol. Mass. | 326.823 |
|---|
| SMILES | CN1CCN(CC1)C1=Nc2cc(Cl)ccc2Nc2ccccc12 |t:8| |
|---|
| Structure | 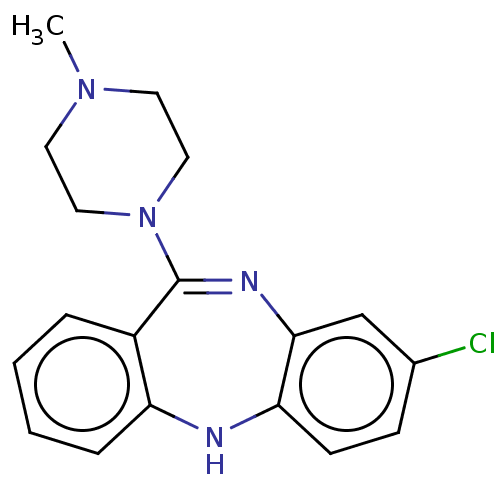
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Liégeois, JF; Bruhwyler, J; Damas, J; Nguyen, TP; Chleide, EM; Mercier, MG; Rogister, FA; Delarge, JE New pyridobenzodiazepine derivatives as potential antipsychotics: synthesis and neurochemical study. J Med Chem36:2107-14 (1993) [PubMed]
Liégeois, JF; Bruhwyler, J; Damas, J; Nguyen, TP; Chleide, EM; Mercier, MG; Rogister, FA; Delarge, JE New pyridobenzodiazepine derivatives as potential antipsychotics: synthesis and neurochemical study. J Med Chem36:2107-14 (1993) [PubMed]