

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Induced myeloid leukemia cell differentiation protein Mcl-1 | ||
| Ligand | BDBM50519059 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1873795 (CHEMBL4375084) | ||
| Ki | 76±n/a nM | ||
| Citation |  Szlávik, Z; Ondi, L; Csékei, M; Paczal, A; Szabó, ZB; Radics, G; Murray, J; Davidson, J; Chen, I; Davis, B; Hubbard, RE; Pedder, C; Dokurno, P; Surgenor, A; Smith, J; Robertson, A; LeToumelin-Braizat, G; Cauquil, N; Zarka, M; Demarles, D; Perron-Sierra, F; Claperon, A; Colland, F; Geneste, O; Kotschy, A Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity. J Med Chem62:6913-6924 (2019) [PubMed] Article Szlávik, Z; Ondi, L; Csékei, M; Paczal, A; Szabó, ZB; Radics, G; Murray, J; Davidson, J; Chen, I; Davis, B; Hubbard, RE; Pedder, C; Dokurno, P; Surgenor, A; Smith, J; Robertson, A; LeToumelin-Braizat, G; Cauquil, N; Zarka, M; Demarles, D; Perron-Sierra, F; Claperon, A; Colland, F; Geneste, O; Kotschy, A Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity. J Med Chem62:6913-6924 (2019) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Induced myeloid leukemia cell differentiation protein Mcl-1 | |||
| Name: | Induced myeloid leukemia cell differentiation protein Mcl-1 | ||
| Synonyms: | BCL2L3 | Bcl-2-like protein 3 | Bcl-2-like protein 3 (Mcl-1) | Bcl-2-related protein EAT/mcl1 | Bcl2-L-3 | Induced myeloid leukemia cell differentiation protein (Mcl-1) | MCL1 | MCL1_HUMAN | Mcl-1 | Myeloid Cell factor-1 (Mcl-1) | Myeloid cell leukemia sequence 1 (BCL2-related) | mcl1/EAT | ||
| Type: | Membrane; Single-pass membrane protein | ||
| Mol. Mass.: | 37332.87 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q07820 | ||
| Residue: | 350 | ||
| Sequence: |
| ||
| BDBM50519059 | |||
| n/a | |||
| Name | BDBM50519059 | ||
| Synonyms: | CHEMBL4573959 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C29H28ClN3O4S | ||
| Mol. Mass. | 550.068 | ||
| SMILES | CCc1sc2ncnc(O[C@H](Cc3ccccc3)C(O)=O)c2c1-c1cc2cc(CN(C)C)oc2c(Cl)c1C |r,wU:10.10,(65.57,-17.15,;64.8,-15.82,;63.26,-15.82,;62.36,-17.06,;60.89,-16.59,;59.56,-17.37,;58.21,-16.6,;58.22,-15.05,;59.55,-14.28,;59.55,-12.74,;58.22,-11.97,;58.21,-10.43,;56.88,-9.67,;56.89,-8.13,;55.55,-7.36,;54.22,-8.13,;54.22,-9.68,;55.56,-10.44,;56.88,-12.75,;55.55,-11.98,;56.89,-14.29,;60.89,-15.05,;62.36,-14.57,;62.82,-13.1,;61.8,-11.97,;62.26,-10.51,;61.48,-9.17,;62.51,-8.01,;62.18,-6.51,;63.32,-5.47,;62.99,-3.97,;64.79,-5.94,;63.93,-8.64,;63.78,-10.18,;64.81,-11.32,;66.31,-10.99,;64.34,-12.79,;65.37,-13.93,)| | ||
| Structure | 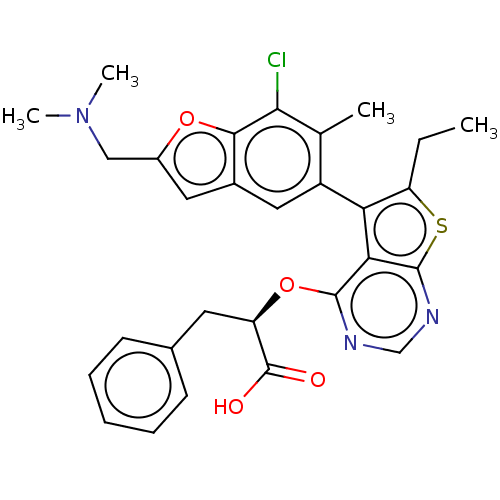
| ||