

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Amyloid-beta precursor protein | ||
| Ligand | BDBM50323202 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1888363 (CHEMBL4390040) | ||
| IC50 | >50000±n/a nM | ||
| Citation |  Sirimangkalakitti, N; Juliawaty, LD; Hakim, EH; Waliana, I; Saito, N; Koyama, K; Kinoshita, K Naturally occurring biflavonoids with amyloid ? aggregation inhibitory activity for development of anti-Alzheimer agents. Bioorg Med Chem Lett29:1994-1997 (2019) [PubMed] Article Sirimangkalakitti, N; Juliawaty, LD; Hakim, EH; Waliana, I; Saito, N; Koyama, K; Kinoshita, K Naturally occurring biflavonoids with amyloid ? aggregation inhibitory activity for development of anti-Alzheimer agents. Bioorg Med Chem Lett29:1994-1997 (2019) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Amyloid-beta precursor protein | |||
| Name: | Amyloid-beta precursor protein | ||
| Synonyms: | A4 | A4_HUMAN | ABPP | AD1 | AICD-50 | AICD-57 | AICD-59 | AID(50) | AID(57) | AID(59) | APP | APPI | Alzheimer disease amyloid protein | Amyloid beta A4 protein | Amyloid beta Protein | Amyloid beta protein (sAPPbeta) | Amyloid beta protein Abeta(1-42) | Amyloid intracellular domain 50 | Amyloid intracellular domain 57 | Amyloid intracellular domain 59 | Amyloid protein (Abeta42b) | Amyloid β-protein (Aβ42) | Beta amyloid A4 protein | Beta-APP40 | Beta-APP42 | Beta-amyloid protein 40 | Beta-amyloid protein 42 | C31 | C83 | C99 | CVAP | Cerebral vascular amyloid peptide | Gamma Secretase | Gamma-CTF(50) | Gamma-CTF(57) | Gamma-CTF(59) | Gamma-secretase | Gamma-secretase C-terminal fragment 50 | Gamma-secretase C-terminal fragment 57 | Gamma-secretase C-terminal fragment 59 | P3(40) | P3(42) | PN-II | PreA4 | Protease nexin-II | S-APP-alpha | S-APP-beta | Soluble APP-alpha | Soluble APP-beta | beta-Amyloid Precursor Protein (APP) | ||
| Type: | Single-pass type I membrane protein | ||
| Mol. Mass.: | 86890.41 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P05067 | ||
| Residue: | 770 | ||
| Sequence: |
| ||
| BDBM50323202 | |||
| n/a | |||
| Name | BDBM50323202 | ||
| Synonyms: | 4',7''-O-methylamentoflavone | 4',7''-di-O-methyl-amentoflavone | 4',7''-di-O-methylamentoflavone | CHEMBL208988 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C32H22O10 | ||
| Mol. Mass. | 566.5111 | ||
| SMILES | COc1ccc(cc1-c1c(OC)cc(O)c2c1oc(cc2=O)-c1ccc(O)cc1)-c1cc(O)c2c(cc(O)cc2=O)o1 |(3.11,3.19,;1.77,3.96,;.44,3.18,;-.9,3.95,;-2.23,3.17,;-2.22,1.64,;-.88,.87,;.45,1.64,;1.63,-.31,;.31,-1.08,;-1.02,-.31,;-2.35,-1.08,;.31,-2.62,;1.63,-3.39,;1.63,-4.93,;2.97,-2.62,;2.97,-1.07,;4.3,-.3,;5.64,-1.08,;5.64,-2.62,;4.3,-3.39,;4.3,-4.93,;6.98,-.31,;8.3,-1.08,;9.64,-.32,;9.64,1.22,;10.97,1.99,;8.3,1.99,;6.98,1.22,;-3.55,.87,;-3.54,-.68,;-4.87,-1.45,;-4.87,-2.99,;-6.21,-.69,;-6.22,.85,;-7.56,1.62,;-8.88,.85,;-10.21,1.62,;-8.88,-.7,;-7.55,-1.47,;-7.56,-3,;-4.89,1.64,)| | ||
| Structure | 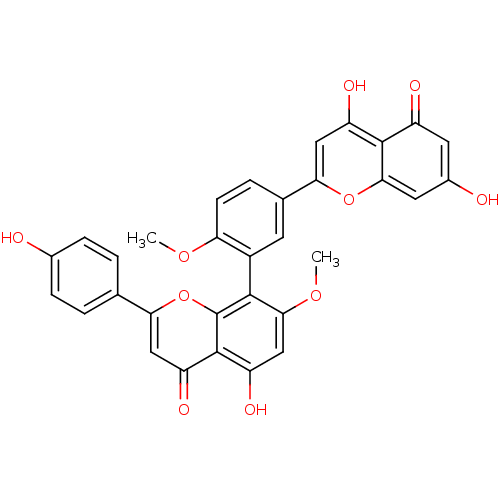
| ||