| Reaction Details |
|---|
 | Report a problem with these data |
| Target | NAD-dependent protein deacylase sirtuin-6 |
|---|
| Ligand | BDBM50528216 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1904594 (CHEMBL4406952) |
|---|
| EC50 | 5700±n/a nM |
|---|
| Citation |  Lu, S; He, X; Ni, D; Zhang, J Allosteric Modulator Discovery: From Serendipity to Structure-Based Design. J Med Chem62:6405-6421 (2019) [PubMed] Article Lu, S; He, X; Ni, D; Zhang, J Allosteric Modulator Discovery: From Serendipity to Structure-Based Design. J Med Chem62:6405-6421 (2019) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| NAD-dependent protein deacylase sirtuin-6 |
|---|
| Name: | NAD-dependent protein deacylase sirtuin-6 |
|---|
| Synonyms: | NAD-dependent protein deacetylase sirtuin-6 | NAD-dependent protein deacetylase sirtuin-6 (SIRT6) | Regulatory protein SIR2 homolog 6 | SIR2-like protein 6 | SIR2L6 | SIR6_HUMAN | SIRT6 | Sirtuin-6 (SIRT6) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 39133.03 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q8N6T7 |
|---|
| Residue: | 355 |
|---|
| Sequence: | MSVNYAAGLSPYADKGKCGLPEIFDPPEELERKVWELARLVWQSSSVVFHTGAGISTASG
IPDFRGPHGVWTMEERGLAPKFDTTFESARPTQTHMALVQLERVGLLRFLVSQNVDGLHV
RSGFPRDKLAELHGNMFVEECAKCKTQYVRDTVVGTMGLKATGRLCTVAKARGLRACRGE
LRDTILDWEDSLPDRDLALADEASRNADLSITLGTSLQIRPSGNLPLATKRRGGRLVIVN
LQPTKHDRHADLRIHGYVDEVMTRLMKHLGLEIPAWDGPRVLERALPPLPRPPTPKLEPK
EESPTRINGSIPAGPKQEPCAQHNGSEPASPKRERPTSPAPHRPPKRVKAKAVPS
|
|
|
|---|
| BDBM50528216 |
|---|
| n/a |
|---|
| Name | BDBM50528216 |
|---|
| Synonyms: | CHEMBL4440358 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H14BrCl2FN2O6S2 |
|---|
| Mol. Mass. | 612.273 |
|---|
| SMILES | Cc1cc(F)c(Br)cc1NS(=O)(=O)c1ccc(NS(=O)(=O)c2cc(Cl)cc(Cl)c2)cc1C(O)=O |
|---|
| Structure | 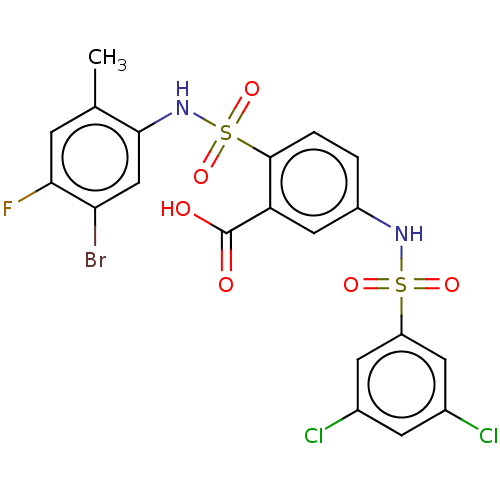
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lu, S; He, X; Ni, D; Zhang, J Allosteric Modulator Discovery: From Serendipity to Structure-Based Design. J Med Chem62:6405-6421 (2019) [PubMed] Article
Lu, S; He, X; Ni, D; Zhang, J Allosteric Modulator Discovery: From Serendipity to Structure-Based Design. J Med Chem62:6405-6421 (2019) [PubMed] Article