| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Heat shock protein HSP 90-alpha |
|---|
| Ligand | BDBM50555444 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2051997 (CHEMBL4706998) |
|---|
| IC50 | 29±n/a nM |
|---|
| Citation |  Jiang, F; Wang, HJ; Jin, YH; Zhang, Q; Wang, ZH; Jia, JM; Liu, F; Wang, L; Bao, QC; Li, DD; You, QD; Xu, XL Novel Tetrahydropyrido[4,3-d]pyrimidines as Potent Inhibitors of Chaperone Heat Shock Protein 90. J Med Chem59:10498-10519 (2016) [PubMed] Jiang, F; Wang, HJ; Jin, YH; Zhang, Q; Wang, ZH; Jia, JM; Liu, F; Wang, L; Bao, QC; Li, DD; You, QD; Xu, XL Novel Tetrahydropyrido[4,3-d]pyrimidines as Potent Inhibitors of Chaperone Heat Shock Protein 90. J Med Chem59:10498-10519 (2016) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Heat shock protein HSP 90-alpha |
|---|
| Name: | Heat shock protein HSP 90-alpha |
|---|
| Synonyms: | HS90A_HUMAN | HSP 86 | HSP86 | HSP90A | HSP90AA1 | HSPC1 | HSPCA | Heat Shock Protein 90 (Hsp90) | Heat shock 86 kDa | Heat shock protein HSP 90 (HSP90) | Heat shock protein HSP 90-alpha (HSP90) | Heat shock protein HSP 90-alpha (HSP90A) | LAP-2 | LPS-associated protein 2 | Lipopolysaccharide-associated protein 2 | Renal carcinoma antigen NY-REN-38 | heat shock protein 90kDa alpha (cytosolic), class A member 1 isoform 2 |
|---|
| Type: | Molecular Chaperone |
|---|
| Mol. Mass.: | 84623.45 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P07900 |
|---|
| Residue: | 732 |
|---|
| Sequence: | MPEETQTQDQPMEEEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIR
YESLTDPSKLDSGKELHINLIPNKQDRTLTIVDTGIGMTKADLINNLGTIAKSGTKAFME
ALQAGADISMIGQFGVGFYSAYLVAEKVTVITKHNDDEQYAWESSAGGSFTVRTDTGEPM
GRGTKVILHLKEDQTEYLEERRIKEIVKKHSQFIGYPITLFVEKERDKEVSDDEAEEKED
KEEEKEKEEKESEDKPEIEDVGSDEEEEKKDGDKKKKKKIKEKYIDQEELNKTKPIWTRN
PDDITNEEYGEFYKSLTNDWEDHLAVKHFSVEGQLEFRALLFVPRRAPFDLFENRKKKNN
IKLYVRRVFIMDNCEELIPEYLNFIRGVVDSEDLPLNISREMLQQSKILKVIRKNLVKKC
LELFTELAEDKENYKKFYEQFSKNIKLGIHEDSQNRKKLSELLRYYTSASGDEMVSLKDY
CTRMKENQKHIYYITGETKDQVANSAFVERLRKHGLEVIYMIEPIDEYCVQQLKEFEGKT
LVSVTKEGLELPEDEEEKKKQEEKKTKFENLCKIMKDILEKKVEKVVVSNRLVTSPCCIV
TSTYGWTANMERIMKAQALRDNSTMGYMAAKKHLEINPDHSIIETLRQKAEADKNDKSVK
DLVILLYETALLSSGFSLEDPQTHANRIYRMIKLGLGIDEDDPTADDTSAAVTEEMPPLE
GDDDTSRMEEVD
|
|
|
|---|
| BDBM50555444 |
|---|
| n/a |
|---|
| Name | BDBM50555444 |
|---|
| Synonyms: | CHEMBL4746663 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H30N4O4 |
|---|
| Mol. Mass. | 426.5087 |
|---|
| SMILES | CC(C)c1cc(C(=O)N2CCc3nc(OCCN4CCCC4)ncc3C2)c(O)cc1O |
|---|
| Structure | 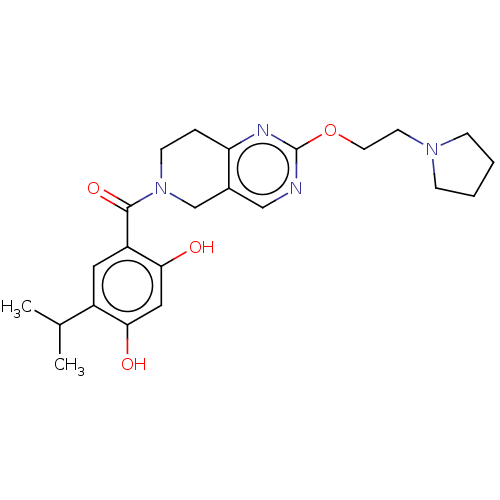
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Jiang, F; Wang, HJ; Jin, YH; Zhang, Q; Wang, ZH; Jia, JM; Liu, F; Wang, L; Bao, QC; Li, DD; You, QD; Xu, XL Novel Tetrahydropyrido[4,3-d]pyrimidines as Potent Inhibitors of Chaperone Heat Shock Protein 90. J Med Chem59:10498-10519 (2016) [PubMed]
Jiang, F; Wang, HJ; Jin, YH; Zhang, Q; Wang, ZH; Jia, JM; Liu, F; Wang, L; Bao, QC; Li, DD; You, QD; Xu, XL Novel Tetrahydropyrido[4,3-d]pyrimidines as Potent Inhibitors of Chaperone Heat Shock Protein 90. J Med Chem59:10498-10519 (2016) [PubMed]