| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Dual specificity tyrosine-phosphorylation-regulated kinase 1A |
|---|
| Ligand | BDBM50560703 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2072743 (CHEMBL4728277) |
|---|
| Kd | 100±n/a nM |
|---|
| Citation |  Schröder, M; Bullock, AN; Fedorov, O; Bracher, F; Chaikuad, A; Knapp, S DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity. J Med Chem63:10224-10234 (2020) [PubMed] Article Schröder, M; Bullock, AN; Fedorov, O; Bracher, F; Chaikuad, A; Knapp, S DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity. J Med Chem63:10224-10234 (2020) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Dual specificity tyrosine-phosphorylation-regulated kinase 1A |
|---|
| Name: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A |
|---|
| Synonyms: | DYR1A_HUMAN | DYRK | DYRK1A | Dual specificity YAK1-related kinase | Dual specificity YAK1-related kinase 1A (Dyrk1A) | Dual specificity tyrosine-phosphorylation-regulated kinase 1A (DYRK1A) | Dual-specificity tyrosine-phosphorylation regulated kinase 1A | Dual-specificity tyrosine-regulated kinases 1A | HP86 | MNB | MNBH |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 85616.61 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q13627 |
|---|
| Residue: | 763 |
|---|
| Sequence: | MHTGGETSACKPSSVRLAPSFSFHAAGLQMAGQMPHSHQYSDRRQPNISDQQVSALSYSD
QIQQPLTNQVMPDIVMLQRRMPQTFRDPATAPLRKLSVDLIKTYKHINEVYYAKKKRRHQ
QGQGDDSSHKKERKVYNDGYDDDNYDYIVKNGEKWMDRYEIDSLIGKGSFGQVVKAYDRV
EQEWVAIKIIKNKKAFLNQAQIEVRLLELMNKHDTEMKYYIVHLKRHFMFRNHLCLVFEM
LSYNLYDLLRNTNFRGVSLNLTRKFAQQMCTALLFLATPELSIIHCDLKPENILLCNPKR
SAIKIVDFGSSCQLGQRIYQYIQSRFYRSPEVLLGMPYDLAIDMWSLGCILVEMHTGEPL
FSGANEVDQMNKIVEVLGIPPAHILDQAPKARKFFEKLPDGTWNLKKTKDGKREYKPPGT
RKLHNILGVETGGPGGRRAGESGHTVADYLKFKDLILRMLDYDPKTRIQPYYALQHSFFK
KTADEGTNTSNSVSTSPAMEQSQSSGTTSSTSSSSGGSSGTSNSGRARSDPTHQHRHSGG
HFTAAVQAMDCETHSPQVRQQFPAPLGWSGTEAPTQVTVETHPVQETTFHVAPQQNALHH
HHGNSSHHHHHHHHHHHHHGQQALGNRTRPRVYNSPTNSSSTQDSMEVGHSHHSMTSLSS
STTSSSTSSSSTGNQGNQAYQNRPVAANTLDFGQNGAMDVNLTVYSNPRQETGIAGHPTY
QFSANTGPAHYMTEGHLTMRQGADREESPMTGVCVQQSPVASS
|
|
|
|---|
| BDBM50560703 |
|---|
| n/a |
|---|
| Name | BDBM50560703 |
|---|
| Synonyms: | CHEMBL1349996 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H15NO2S |
|---|
| Mol. Mass. | 249.329 |
|---|
| SMILES | CCN1\C(Sc2ccc(OC)cc12)=C/C(C)=O |
|---|
| Structure | 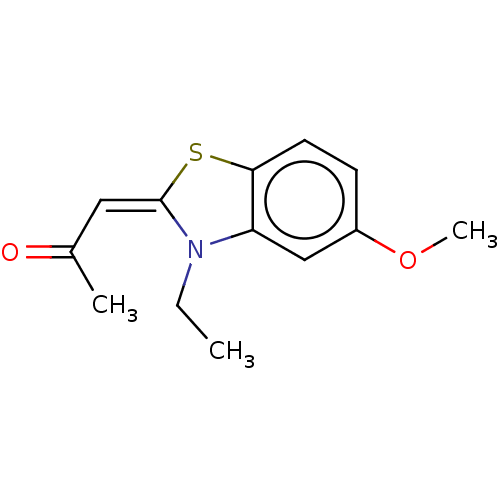
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Schröder, M; Bullock, AN; Fedorov, O; Bracher, F; Chaikuad, A; Knapp, S DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity. J Med Chem63:10224-10234 (2020) [PubMed] Article
Schröder, M; Bullock, AN; Fedorov, O; Bracher, F; Chaikuad, A; Knapp, S DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity. J Med Chem63:10224-10234 (2020) [PubMed] Article