

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Activin receptor type-1 | ||
| Ligand | BDBM50568934 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_2107053 (CHEMBL4815728) | ||
| IC50 | 92±n/a nM | ||
| Citation |  Nikhar, S; Siokas, I; Schlicher, L; Lee, S; Gyrd-Hansen, M; Degterev, A; Cuny, GD Design of pyrido[2,3-d]pyrimidin-7-one inhibitors of receptor interacting protein kinase-2 (RIPK2) and nucleotide-binding oligomerization domain (NOD) cell signaling. Eur J Med Chem215:0 (2021) [PubMed] Article Nikhar, S; Siokas, I; Schlicher, L; Lee, S; Gyrd-Hansen, M; Degterev, A; Cuny, GD Design of pyrido[2,3-d]pyrimidin-7-one inhibitors of receptor interacting protein kinase-2 (RIPK2) and nucleotide-binding oligomerization domain (NOD) cell signaling. Eur J Med Chem215:0 (2021) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Activin receptor type-1 | |||
| Name: | Activin receptor type-1 | ||
| Synonyms: | 2.7.11.30 | ACTR-I | ACVR1 | ACVR1_HUMAN | ACVRLK2 | ALK-2 | ALK2/ACVR1 | Activin receptor type I | Activin receptor-like kinase 2 | Activin receptor-like kinase 2 (ALK-2) | Activin receptor-like kinase 2 (ALK2/ACVR1) | Q04771 | SKR1 | Serine/threonine-protein kinase receptor R1 | TGF-B superfamily receptor type I | TSR-I | ||
| Type: | n/a | ||
| Mol. Mass.: | 57158.32 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | n/a | ||
| Residue: | 509 | ||
| Sequence: |
| ||
| BDBM50568934 | |||
| n/a | |||
| Name | BDBM50568934 | ||
| Synonyms: | CHEMBL4864576 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C33H33Cl2N5O4S | ||
| Mol. Mass. | 666.617 | ||
| SMILES | CCN(CC)CCOc1ccc(Nc2ncc3cc(-c4c(Cl)cccc4Cl)c(=O)n(Cc4cccc(c4)S(C)(=O)=O)c3n2)cc1 |(26.34,-6.01,;27.68,-5.24,;29.01,-6.01,;29.01,-7.55,;27.68,-8.32,;30.34,-5.24,;31.68,-6,;33.01,-5.23,;34.34,-6,;34.34,-7.55,;35.68,-8.32,;37.01,-7.55,;38.35,-8.32,;39.68,-7.54,;39.67,-6.01,;40.99,-5.23,;42.34,-6,;43.67,-5.23,;45.01,-6.01,;46.35,-5.25,;46.35,-3.71,;45.01,-2.94,;47.68,-2.94,;49.02,-3.72,;49.01,-5.26,;47.67,-6.02,;47.66,-7.56,;45,-7.55,;46.33,-8.33,;43.66,-8.32,;43.65,-9.86,;42.32,-10.62,;40.99,-9.84,;39.66,-10.6,;39.65,-12.14,;40.99,-12.92,;42.32,-12.15,;40.98,-14.45,;39.65,-15.22,;42.47,-14.05,;42.07,-15.54,;42.33,-7.54,;41.01,-8.31,;37.01,-6,;35.67,-5.23,)| | ||
| Structure | 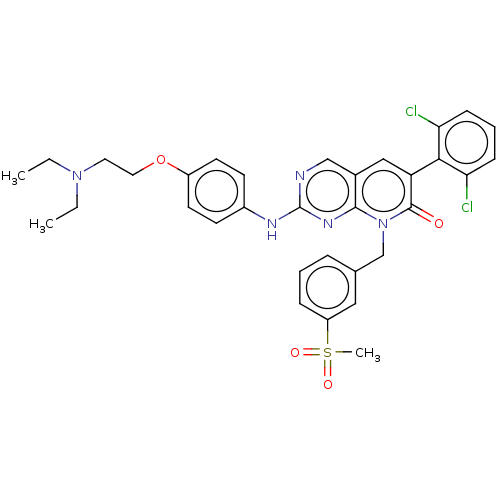
| ||