| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Endoplasmic reticulum aminopeptidase 1 |
|---|
| Ligand | BDBM50570851 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2113192 (CHEMBL4822042) |
|---|
| IC50 | 330±n/a nM |
|---|
| Citation |  Wilding, B; Pasqua, AE; E A Chessum, N; Pierrat, OA; Hahner, T; Tomlin, K; Shehu, E; Burke, R; Richards, GM; Whitton, B; Arwert, EN; Thapaliya, A; Salimraj, R; van Montfort, R; Skawinska, A; Hayes, A; Raynaud, F; Chopra, R; Jones, K; Newton, G; Cheeseman, MD Investigating the phosphinic acid tripeptide mimetic DG013A as a tool compound inhibitor of the M1-aminopeptidase ERAP1. Bioorg Med Chem Lett42:0 (2021) [PubMed] Article Wilding, B; Pasqua, AE; E A Chessum, N; Pierrat, OA; Hahner, T; Tomlin, K; Shehu, E; Burke, R; Richards, GM; Whitton, B; Arwert, EN; Thapaliya, A; Salimraj, R; van Montfort, R; Skawinska, A; Hayes, A; Raynaud, F; Chopra, R; Jones, K; Newton, G; Cheeseman, MD Investigating the phosphinic acid tripeptide mimetic DG013A as a tool compound inhibitor of the M1-aminopeptidase ERAP1. Bioorg Med Chem Lett42:0 (2021) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Endoplasmic reticulum aminopeptidase 1 |
|---|
| Name: | Endoplasmic reticulum aminopeptidase 1 |
|---|
| Synonyms: | APPILS | ARTS1 | Aminopeptidase | ERAP1 | ERAP1_HUMAN | KIAA0525 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 107232.03 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_1473167 |
|---|
| Residue: | 941 |
|---|
| Sequence: | MVFLPLKWSLATMSFLLSSLLALLTVSTPSWCQSTEASPKRSDGTPFPWNKIRLPEYVIP
VHYDLLIHANLTTLTFWGTTKVEITASQPTSTIILHSHHLQISRATLRKGAGERLSEEPL
QVLEHPRQEQIALLAPEPLLVGLPYTVVIHYAGNLSETFHGFYKSTYRTKEGELRILAST
QFEPTAARMAFPCFDEPAFKASFSIKIRREPRHLAISNMPLVKSVTVAEGLIEDHFDVTV
KMSTYLVAFIISDFESVSKITKSGVKVSVYAVPDKINQADYALDAAVTLLEFYEDYFSIP
YPLPKQDLAAIPDFQSGAMENWGLTTYRESALLFDAEKSSASSKLGITMTVAHELAHQWF
GNLVTMEWWNDLWLNEGFAKFMEFVSVSVTHPELKVGDYFFGKCFDAMEVDALNSSHPVS
TPVENPAQIREMFDDVSYDKGACILNMLREYLSADAFKSGIVQYLQKHSYKNTKNEDLWD
SMASICPTDGVKGMDGFCSRSQHSSSSSHWHQEGVDVKTMMNTWTLQKGFPLITITVRGR
NVHMKQEHYMKGSDGAPDTGYLWHVPLTFITSKSDMVHRFLLKTKTDVLILPEEVEWIKF
NVGMNGYYIVHYEDDGWDSLTGLLKGTHTAVSSNDRASLINNAFQLVSIGKLSIEKALDL
SLYLKHETEIMPVFQGLNELIPMYKLMEKRDMNEVETQFKAFLIRLLRDLIDKQTWTDEG
SVSERMLRSQLLLLACVHNYQPCVQRAEGYFRKWKESNGNLSLPVDVTLAVFAVGAQSTE
GWDFLYSKYQFSLSSTEKSQIEFALCRTQNKEKLQWLLDESFKGDKIKTQEFPQILTLIG
RNPVGYPLAWQFLRKNWNKLVQKFELGSSSIAHMVMGTTNQFSTRTRLEEVKGFFSSLKE
NGSQLRCVQQTIETIEENIGWMDKNFDKIRVWLQSEKLERM
|
|
|
|---|
| BDBM50570851 |
|---|
| n/a |
|---|
| Name | BDBM50570851 |
|---|
| Synonyms: | CHEMBL4853049 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H36N3O3P |
|---|
| Mol. Mass. | 469.5561 |
|---|
| SMILES | CC(C)C[C@H](CP(O)(=O)[C@@H](N)CCc1ccccc1)C(=O)NCCc1c[nH]c2ccccc12 |r| |
|---|
| Structure | 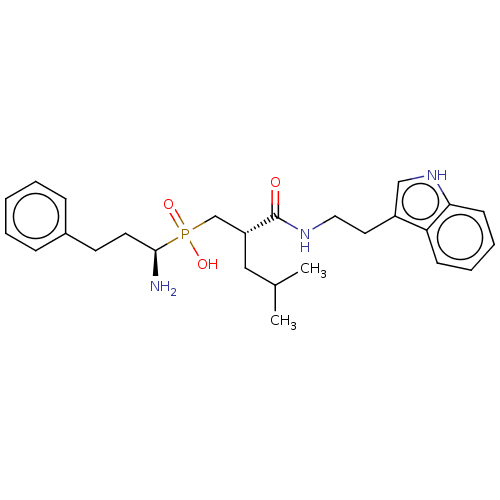
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wilding, B; Pasqua, AE; E A Chessum, N; Pierrat, OA; Hahner, T; Tomlin, K; Shehu, E; Burke, R; Richards, GM; Whitton, B; Arwert, EN; Thapaliya, A; Salimraj, R; van Montfort, R; Skawinska, A; Hayes, A; Raynaud, F; Chopra, R; Jones, K; Newton, G; Cheeseman, MD Investigating the phosphinic acid tripeptide mimetic DG013A as a tool compound inhibitor of the M1-aminopeptidase ERAP1. Bioorg Med Chem Lett42:0 (2021) [PubMed] Article
Wilding, B; Pasqua, AE; E A Chessum, N; Pierrat, OA; Hahner, T; Tomlin, K; Shehu, E; Burke, R; Richards, GM; Whitton, B; Arwert, EN; Thapaliya, A; Salimraj, R; van Montfort, R; Skawinska, A; Hayes, A; Raynaud, F; Chopra, R; Jones, K; Newton, G; Cheeseman, MD Investigating the phosphinic acid tripeptide mimetic DG013A as a tool compound inhibitor of the M1-aminopeptidase ERAP1. Bioorg Med Chem Lett42:0 (2021) [PubMed] Article