| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Fatty-acid amide hydrolase 1 |
|---|
| Ligand | BDBM26739 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2172610 (CHEMBL5057744) |
|---|
| IC50 | 32±n/a nM |
|---|
| Citation |  Wilt, S; Kodani, S; Valencia, L; Hudson, PK; Sanchez, S; Quintana, T; Morisseau, C; Hammock, BD; Kandasamy, R; Pecic, S Further exploration of the structure-activity relationship of dual soluble epoxide hydrolase/fatty acid amide hydrolase inhibitors. Bioorg Med Chem51:0 (2021) [PubMed] Article Wilt, S; Kodani, S; Valencia, L; Hudson, PK; Sanchez, S; Quintana, T; Morisseau, C; Hammock, BD; Kandasamy, R; Pecic, S Further exploration of the structure-activity relationship of dual soluble epoxide hydrolase/fatty acid amide hydrolase inhibitors. Bioorg Med Chem51:0 (2021) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Fatty-acid amide hydrolase 1 |
|---|
| Name: | Fatty-acid amide hydrolase 1 |
|---|
| Synonyms: | Anandamide amidohydrolase | Anandamide amidohydrolase 1 | FAAH | FAAH1 | FAAH1_HUMAN | Fatty Acid Amide Hydrolase (FAAH) | Fatty-acid amide hydrolase (FAAH) | Fatty-acid amide hydrolase 1 | Oleamide hydrolase 1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 63071.19 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O00519 |
|---|
| Residue: | 579 |
|---|
| Sequence: | MVQYELWAALPGASGVALACCFVAAAVALRWSGRRTARGAVVRARQRQRAGLENMDRAAQ
RFRLQNPDLDSEALLALPLPQLVQKLHSRELAPEAVLFTYVGKAWEVNKGTNCVTSYLAD
CETQLSQAPRQGLLYGVPVSLKECFTYKGQDSTLGLSLNEGVPAECDSVVVHVLKLQGAV
PFVHTNVPQSMFSYDCSNPLFGQTVNPWKSSKSPGGSSGGEGALIGSGGSPLGLGTDIGG
SIRFPSSFCGICGLKPTGNRLSKSGLKGCVYGQEAVRLSVGPMARDVESLALCLRALLCE
DMFRLDPTVPPLPFREEVYTSSQPLRVGYYETDNYTMPSPAMRRAVLETKQSLEAAGHTL
VPFLPSNIPHALETLSTGGLFSDGGHTFLQNFKGDFVDPCLGDLVSILKLPQWLKGLLAF
LVKPLLPRLSAFLSNMKSRSAGKLWELQHEIEVYRKTVIAQWRALDLDVVLTPMLAPALD
LNAPGRATGAVSYTMLYNCLDFPAGVVPVTTVTAEDEAQMEHYRGYFGDIWDKMLQKGMK
KSVGLPVAVQCVALPWQEELCLRFMREVERLMTPEKQSS
|
|
|
|---|
| BDBM26739 |
|---|
| n/a |
|---|
| Name | BDBM26739 |
|---|
| Synonyms: | 3-(3-carbamoylphenyl)phenyl N-cyclohexylcarbamate | CHEMBL184238 | URB 597 | URB-597 | URB597 | US9187413, 1a (URB597) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H22N2O3 |
|---|
| Mol. Mass. | 338.4003 |
|---|
| SMILES | NC(=O)c1cccc(c1)-c1cccc(OC(=O)NC2CCCCC2)c1 |
|---|
| Structure | 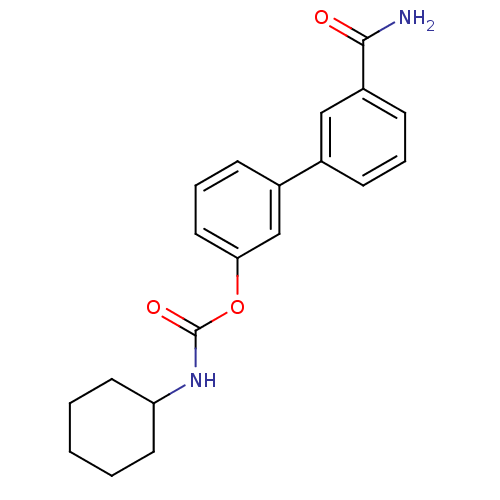
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wilt, S; Kodani, S; Valencia, L; Hudson, PK; Sanchez, S; Quintana, T; Morisseau, C; Hammock, BD; Kandasamy, R; Pecic, S Further exploration of the structure-activity relationship of dual soluble epoxide hydrolase/fatty acid amide hydrolase inhibitors. Bioorg Med Chem51:0 (2021) [PubMed] Article
Wilt, S; Kodani, S; Valencia, L; Hudson, PK; Sanchez, S; Quintana, T; Morisseau, C; Hammock, BD; Kandasamy, R; Pecic, S Further exploration of the structure-activity relationship of dual soluble epoxide hydrolase/fatty acid amide hydrolase inhibitors. Bioorg Med Chem51:0 (2021) [PubMed] Article