

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Activin receptor type-1 | ||
| Ligand | BDBM50593561 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_2209043 (CHEMBL5121992) | ||
| IC50 | 17±n/a nM | ||
| Citation |  Ullrich, T; Arista, L; Weiler, S; Teixeira-Fouchard, S; Broennimann, V; Stiefl, N; Head, V; Kramer, I; Guth, S Discovery of a novel 2-aminopyrazine-3-carboxamide as a potent and selective inhibitor of Activin Receptor-Like Kinase-2 (ALK2) for the treatment of fibrodysplasia ossificans progressiva. Bioorg Med Chem Lett64:0 (2022) [PubMed] Article Ullrich, T; Arista, L; Weiler, S; Teixeira-Fouchard, S; Broennimann, V; Stiefl, N; Head, V; Kramer, I; Guth, S Discovery of a novel 2-aminopyrazine-3-carboxamide as a potent and selective inhibitor of Activin Receptor-Like Kinase-2 (ALK2) for the treatment of fibrodysplasia ossificans progressiva. Bioorg Med Chem Lett64:0 (2022) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Activin receptor type-1 | |||
| Name: | Activin receptor type-1 | ||
| Synonyms: | 2.7.11.30 | ACTR-I | ACVR1 | ACVR1_HUMAN | ACVRLK2 | ALK-2 | ALK2/ACVR1 | Activin receptor type I | Activin receptor-like kinase 2 | Activin receptor-like kinase 2 (ALK-2) | Activin receptor-like kinase 2 (ALK2/ACVR1) | Q04771 | SKR1 | Serine/threonine-protein kinase receptor R1 | TGF-B superfamily receptor type I | TSR-I | ||
| Type: | n/a | ||
| Mol. Mass.: | 57158.32 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | n/a | ||
| Residue: | 509 | ||
| Sequence: |
| ||
| BDBM50593561 | |||
| n/a | |||
| Name | BDBM50593561 | ||
| Synonyms: | CHEMBL5200388 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C27H38N6O2 | ||
| Mol. Mass. | 478.6296 | ||
| SMILES | Nc1ncc(nc1C(=O)N[C@H]1CC[C@H](O)CC1)-c1ccc(CN2CCC(CC2)N2CCCC2)cc1 |r,wU:10.10,wD:13.14,(-7.88,3.85,;-6.55,3.08,;-5.21,3.85,;-3.89,3.08,;-3.89,1.54,;-5.21,.78,;-6.55,1.54,;-7.88,.77,;-9.22,1.54,;-7.88,-.77,;-6.55,-1.54,;-5.21,-.77,;-3.88,-1.54,;-3.88,-3.08,;-2.55,-3.85,;-5.21,-3.85,;-6.55,-3.08,;-2.55,.77,;-2.55,-.77,;-1.21,-1.53,;.11,-.77,;1.44,-1.54,;2.78,-.77,;4.11,-1.54,;5.45,-.77,;5.45,.77,;4.11,1.54,;2.78,.77,;6.78,1.54,;8.19,.92,;9.22,2.06,;8.45,3.39,;6.94,3.07,;.11,.77,;-1.22,1.54,)| | ||
| Structure | 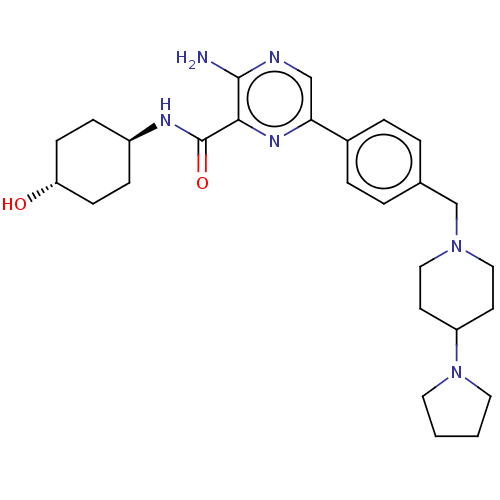
| ||