| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Histone deacetylase 3 |
|---|
| Ligand | BDBM50595953 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2218703 (CHEMBL5132037) |
|---|
| IC50 | 841±n/a nM |
|---|
| Citation |  Toutah, K; Nawar, N; Timonen, S; Sorger, H; Raouf, YS; Bukhari, S; von Jan, J; Ianevski, A; Gawel, JM; Olaoye, OO; Geletu, M; Abdeldayem, A; Israelian, J; Radu, TB; Sedighi, A; Bhatti, MN; Hassan, MM; Manaswiyoungkul, P; Shouksmith, AE; Neubauer, HA; de Araujo, ED; Aittokallio, T; Krämer, OH; Moriggl, R; Mustjoki, S; Herling, M; Gunning, PT Development of HDAC Inhibitors Exhibiting Therapeutic Potential in T-Cell Prolymphocytic Leukemia. J Med Chem64:8486-8509 (2021) [PubMed] Article Toutah, K; Nawar, N; Timonen, S; Sorger, H; Raouf, YS; Bukhari, S; von Jan, J; Ianevski, A; Gawel, JM; Olaoye, OO; Geletu, M; Abdeldayem, A; Israelian, J; Radu, TB; Sedighi, A; Bhatti, MN; Hassan, MM; Manaswiyoungkul, P; Shouksmith, AE; Neubauer, HA; de Araujo, ED; Aittokallio, T; Krämer, OH; Moriggl, R; Mustjoki, S; Herling, M; Gunning, PT Development of HDAC Inhibitors Exhibiting Therapeutic Potential in T-Cell Prolymphocytic Leukemia. J Med Chem64:8486-8509 (2021) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Histone deacetylase 3 |
|---|
| Name: | Histone deacetylase 3 |
|---|
| Synonyms: | HD3 | HDAC3 | HDAC3_HUMAN | Histone deacetylase 3 (HDAC3) | Human HDAC3 | RPD3-2 | SMAP45 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 48829.55 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O15379 |
|---|
| Residue: | 428 |
|---|
| Sequence: | MAKTVAYFYDPDVGNFHYGAGHPMKPHRLALTHSLVLHYGLYKKMIVFKPYQASQHDMCR
FHSEDYIDFLQRVSPTNMQGFTKSLNAFNVGDDCPVFPGLFEFCSRYTGASLQGATQLNN
KICDIAINWAGGLHHAKKFEASGFCYVNDIVIGILELLKYHPRVLYIDIDIHHGDGVQEA
FYLTDRVMTVSFHKYGNYFFPGTGDMYEVGAESGRYYCLNVPLRDGIDDQSYKHLFQPVI
NQVVDFYQPTCIVLQCGADSLGCDRLGCFNLSIRGHGECVEYVKSFNIPLLVLGGGGYTV
RNVARCWTYETSLLVEEAISEELPYSEYFEYFAPDFTLHPDVSTRIENQNSRQYLDQIRQ
TIFENLKMLNHAPSVQIHDVPADLLTYDRTDEADAEERGPEENYSRPEAPNEFYDGDHDN
DKESDVEI
|
|
|
|---|
| BDBM50595953 |
|---|
| n/a |
|---|
| Name | BDBM50595953 |
|---|
| Synonyms: | CHEMBL5193014 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H17FN2O4S |
|---|
| Mol. Mass. | 364.391 |
|---|
| SMILES | ONC(=O)c1ccc(CN(C2CC2)S(=O)(=O)c2ccc(F)cc2)cc1 |
|---|
| Structure | 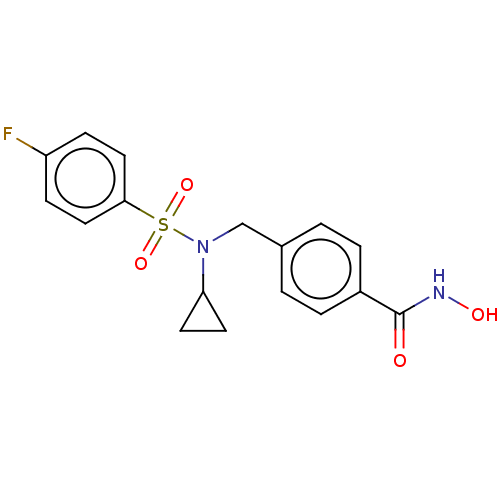
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Toutah, K; Nawar, N; Timonen, S; Sorger, H; Raouf, YS; Bukhari, S; von Jan, J; Ianevski, A; Gawel, JM; Olaoye, OO; Geletu, M; Abdeldayem, A; Israelian, J; Radu, TB; Sedighi, A; Bhatti, MN; Hassan, MM; Manaswiyoungkul, P; Shouksmith, AE; Neubauer, HA; de Araujo, ED; Aittokallio, T; Krämer, OH; Moriggl, R; Mustjoki, S; Herling, M; Gunning, PT Development of HDAC Inhibitors Exhibiting Therapeutic Potential in T-Cell Prolymphocytic Leukemia. J Med Chem64:8486-8509 (2021) [PubMed] Article
Toutah, K; Nawar, N; Timonen, S; Sorger, H; Raouf, YS; Bukhari, S; von Jan, J; Ianevski, A; Gawel, JM; Olaoye, OO; Geletu, M; Abdeldayem, A; Israelian, J; Radu, TB; Sedighi, A; Bhatti, MN; Hassan, MM; Manaswiyoungkul, P; Shouksmith, AE; Neubauer, HA; de Araujo, ED; Aittokallio, T; Krämer, OH; Moriggl, R; Mustjoki, S; Herling, M; Gunning, PT Development of HDAC Inhibitors Exhibiting Therapeutic Potential in T-Cell Prolymphocytic Leukemia. J Med Chem64:8486-8509 (2021) [PubMed] Article