| Reaction Details |
|---|
 | Report a problem with these data |
| Target | NAD-dependent protein deacetylase sirtuin-2 |
|---|
| Ligand | BDBM50178769 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_325279 (CHEMBL859690) |
|---|
| IC50 | 19600±n/a nM |
|---|
| Citation |  Napper, AD; Hixon, J; McDonagh, T; Keavey, K; Pons, JF; Barker, J; Yau, WT; Amouzegh, P; Flegg, A; Hamelin, E; Thomas, RJ; Kates, M; Jones, S; Navia, MA; Saunders, JO; DiStefano, PS; Curtis, R Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J Med Chem48:8045-54 (2005) [PubMed] Article Napper, AD; Hixon, J; McDonagh, T; Keavey, K; Pons, JF; Barker, J; Yau, WT; Amouzegh, P; Flegg, A; Hamelin, E; Thomas, RJ; Kates, M; Jones, S; Navia, MA; Saunders, JO; DiStefano, PS; Curtis, R Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J Med Chem48:8045-54 (2005) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| NAD-dependent protein deacetylase sirtuin-2 |
|---|
| Name: | NAD-dependent protein deacetylase sirtuin-2 |
|---|
| Synonyms: | NAD-Dependent Deacetylase Sirtuin-2 | NAD-dependent deacetylase sirtuin 2 | NAD-dependent deacetylase sirtuin 3 | NAD-dependent protein deacetylase sirtuin-2 (SIRT2) | SIR2-like | SIR2-like protein 2 | SIR2L | SIR2L2 | SIR2_HUMAN | SIRT2 | Sirtuin 2 (SIRT2) |
|---|
| Type: | Hydrolase |
|---|
| Mol. Mass.: | 43172.62 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 389 |
|---|
| Sequence: | MAEPDPSHPLETQAGKVQEAQDSDSDSEGGAAGGEADMDFLRNLFSQTLSLGSQKERLLD
ELTLEGVARYMQSERCRRVICLVGAGISTSAGIPDFRSPSTGLYDNLEKYHLPYPEAIFE
ISYFKKHPEPFFALAKELYPGQFKPTICHYFMRLLKDKGLLLRCYTQNIDTLERIAGLEQ
EDLVEAHGTFYTSHCVSASCRHEYPLSWMKEKIFSEVTPKCEDCQSLVKPDIVFFGESLP
ARFFSCMQSDFLKVDLLLVMGTSLQVQPFASLISKAPLSTPRLLINKEKAGQSDPFLGMI
MGLGGGMDFDSKKAYRDVAWLGECDQGCLALAELLGWKKELEDLVRREHASIDAQSGAGV
PNPSTSASPKKSPPPAKDEARTTEREKPQ
|
|
|
|---|
| BDBM50178769 |
|---|
| n/a |
|---|
| Name | BDBM50178769 |
|---|
| Synonyms: | (+/-)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxamide | 6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxamide | CHEMBL420311 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H13ClN2O |
|---|
| Mol. Mass. | 248.708 |
|---|
| SMILES | NC(=O)C1CCCc2c1[nH]c1ccc(Cl)cc21 |
|---|
| Structure | 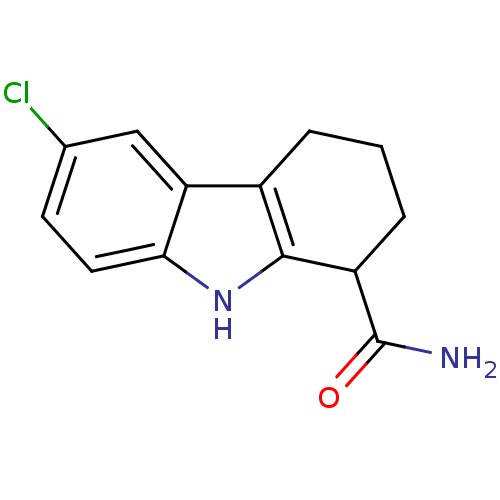
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Napper, AD; Hixon, J; McDonagh, T; Keavey, K; Pons, JF; Barker, J; Yau, WT; Amouzegh, P; Flegg, A; Hamelin, E; Thomas, RJ; Kates, M; Jones, S; Navia, MA; Saunders, JO; DiStefano, PS; Curtis, R Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J Med Chem48:8045-54 (2005) [PubMed] Article
Napper, AD; Hixon, J; McDonagh, T; Keavey, K; Pons, JF; Barker, J; Yau, WT; Amouzegh, P; Flegg, A; Hamelin, E; Thomas, RJ; Kates, M; Jones, S; Navia, MA; Saunders, JO; DiStefano, PS; Curtis, R Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J Med Chem48:8045-54 (2005) [PubMed] Article