| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peroxisome proliferator-activated receptor gamma |
|---|
| Ligand | BDBM50239187 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2244007 (CHEMBL5158217) |
|---|
| EC50 | 54000±n/a nM |
|---|
| Citation |  Sahin, C; Magomedova, L; Ferreira, TAM; Liu, J; Tiefenbach, J; Alves, PS; Queiroz, FJG; Oliveira, AS; Bhattacharyya, M; Grouleff, J; Nogueira, PCN; Silveira, ER; Moreira, DC; Leite, JRSA; Brand, GD; Uehling, D; Poda, G; Krause, H; Cummins, CL; Romeiro, LAS Phenolic Lipids Derived from Cashew Nut Shell Liquid to Treat Metabolic Diseases. J Med Chem65:1961-1978 (2022) [PubMed] Article Sahin, C; Magomedova, L; Ferreira, TAM; Liu, J; Tiefenbach, J; Alves, PS; Queiroz, FJG; Oliveira, AS; Bhattacharyya, M; Grouleff, J; Nogueira, PCN; Silveira, ER; Moreira, DC; Leite, JRSA; Brand, GD; Uehling, D; Poda, G; Krause, H; Cummins, CL; Romeiro, LAS Phenolic Lipids Derived from Cashew Nut Shell Liquid to Treat Metabolic Diseases. J Med Chem65:1961-1978 (2022) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Peroxisome proliferator-activated receptor gamma |
|---|
| Name: | Peroxisome proliferator-activated receptor gamma |
|---|
| Synonyms: | NR1C3 | Nuclear receptor subfamily 1 group C member 3 | PPAR-gamma | PPARG | PPARG_HUMAN | Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor gamma (PPAR gamma) | Peroxisome proliferator-activated receptor gamma (PPARG) | Peroxisome proliferator-activated receptor gamma (PPARγ) | Peroxisome proliferator-activated receptor gamma/Nuclear receptor corepressor 2 | peroxisome proliferator-activated receptor gamma isoform 2 |
|---|
| Type: | Nuclear Receptor |
|---|
| Mol. Mass.: | 57613.46 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P37231 |
|---|
| Residue: | 505 |
|---|
| Sequence: | MGETLGDSPIDPESDSFTDTLSANISQEMTMVDTEMPFWPTNFGISSVDLSVMEDHSHSF
DIKPFTTVDFSSISTPHYEDIPFTRTDPVVADYKYDLKLQEYQSAIKVEPASPPYYSEKT
QLYNKPHEEPSNSLMAIECRVCGDKASGFHYGVHACEGCKGFFRRTIRLKLIYDRCDLNC
RIHKKSRNKCQYCRFQKCLAVGMSHNAIRFGRMPQAEKEKLLAEISSDIDQLNPESADLR
ALAKHLYDSYIKSFPLTKAKARAILTGKTTDKSPFVIYDMNSLMMGEDKIKFKHITPLQE
QSKEVAIRIFQGCQFRSVEAVQEITEYAKSIPGFVNLDLNDQVTLLKYGVHEIIYTMLAS
LMNKDGVLISEGQGFMTREFLKSLRKPFGDFMEPKFEFAVKFNALELDDSDLAIFIAVII
LSGDRPGLLNVKPIEDIQDNLLQALELQLKLNHPESSQLFAKLLQKMTDLRQIVTEHVQL
LQVIKKTETDMSLHPLLQEIYKDLY
|
|
|
|---|
| BDBM50239187 |
|---|
| n/a |
|---|
| Name | BDBM50239187 |
|---|
| Synonyms: | CHEBI:30813 | Decanoic Acid | Decanoic Acid Anion |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C10H20O2 |
|---|
| Mol. Mass. | 172.2646 |
|---|
| SMILES | CCCCCCCCCC(O)=O |
|---|
| Structure | 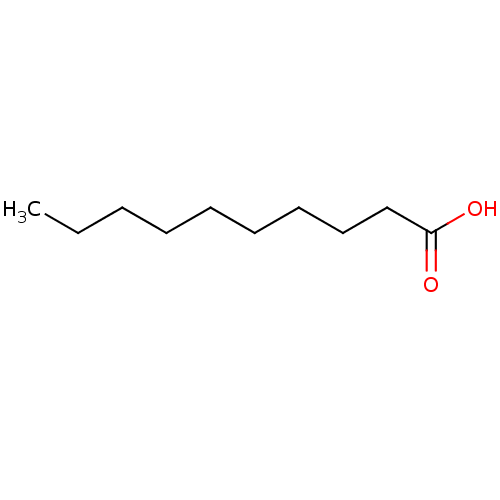
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Sahin, C; Magomedova, L; Ferreira, TAM; Liu, J; Tiefenbach, J; Alves, PS; Queiroz, FJG; Oliveira, AS; Bhattacharyya, M; Grouleff, J; Nogueira, PCN; Silveira, ER; Moreira, DC; Leite, JRSA; Brand, GD; Uehling, D; Poda, G; Krause, H; Cummins, CL; Romeiro, LAS Phenolic Lipids Derived from Cashew Nut Shell Liquid to Treat Metabolic Diseases. J Med Chem65:1961-1978 (2022) [PubMed] Article
Sahin, C; Magomedova, L; Ferreira, TAM; Liu, J; Tiefenbach, J; Alves, PS; Queiroz, FJG; Oliveira, AS; Bhattacharyya, M; Grouleff, J; Nogueira, PCN; Silveira, ER; Moreira, DC; Leite, JRSA; Brand, GD; Uehling, D; Poda, G; Krause, H; Cummins, CL; Romeiro, LAS Phenolic Lipids Derived from Cashew Nut Shell Liquid to Treat Metabolic Diseases. J Med Chem65:1961-1978 (2022) [PubMed] Article