| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cathepsin D |
|---|
| Ligand | BDBM50212178 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_455664 (CHEMBL886447) |
|---|
| IC50 | 275000±n/a nM |
|---|
| Citation |  Lindsley, SR; Moore, KP; Rajapakse, HA; Selnick, HG; Young, MB; Zhu, H; Munshi, S; Kuo, L; McGaughey, GB; Colussi, D; Crouthamel, MC; Lai, MT; Pietrak, B; Price, EA; Sankaranarayanan, S; Simon, AJ; Seabrook, GR; Hazuda, DJ; Pudvah, NT; Hochman, JH; Graham, SL; Vacca, JP; Nantermet, PG Design, synthesis, and SAR of macrocyclic tertiary carbinamine BACE-1 inhibitors. Bioorg Med Chem Lett17:4057-61 (2007) [PubMed] Article Lindsley, SR; Moore, KP; Rajapakse, HA; Selnick, HG; Young, MB; Zhu, H; Munshi, S; Kuo, L; McGaughey, GB; Colussi, D; Crouthamel, MC; Lai, MT; Pietrak, B; Price, EA; Sankaranarayanan, S; Simon, AJ; Seabrook, GR; Hazuda, DJ; Pudvah, NT; Hochman, JH; Graham, SL; Vacca, JP; Nantermet, PG Design, synthesis, and SAR of macrocyclic tertiary carbinamine BACE-1 inhibitors. Bioorg Med Chem Lett17:4057-61 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cathepsin D |
|---|
| Name: | Cathepsin D |
|---|
| Synonyms: | CATD_HUMAN | CPSD | CTSD | Cathepsin D [Precursor] | Cathepsin D heavy chain | Cathepsin D light chain | Cathepsin D precursor |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 44551.72 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Human proCathepsin D (SwissProt accession number P07339) was expressed in Sf9 cells, purified, and autoactivated. |
|---|
| Residue: | 412 |
|---|
| Sequence: | MQPSSLLPLALCLLAAPASALVRIPLHKFTSIRRTMSEVGGSVEDLIAKGPVSKYSQAVP
AVTEGPIPEVLKNYMDAQYYGEIGIGTPPQCFTVVFDTGSSNLWVPSIHCKLLDIACWIH
HKYNSDKSSTYVKNGTSFDIHYGSGSLSGYLSQDTVSVPCQSASSASALGGVKVERQVFG
EATKQPGITFIAAKFDGILGMAYPRISVNNVLPVFDNLMQQKLVDQNIFSFYLSRDPDAQ
PGGELMLGGTDSKYYKGSLSYLNVTRKAYWQVHLDQVEVASGLTLCKEGCEAIVDTGTSL
MVGPVDEVRELQKAIGAVPLIQGEYMIPCEKVSTLPAITLKLGGKGYKLSPEDYTLKVSQ
AGKTLCLSGFMGMDIPPPSGPLWILGDVFIGRYYTVFDRDNNRVGFAEAARL
|
|
|
|---|
| BDBM50212178 |
|---|
| n/a |
|---|
| Name | BDBM50212178 |
|---|
| Synonyms: | CHEMBL248699 | N-((5R,14R)-5-amino-5-methyl-16-oxo-14-phenyl-3-oxa-15-aza-tricyclo[15.3.1.1*7,11*]docosa-1(21),7,9,11(22),17,19-hexaen-19-yl)-N-propyl-methanesulfonamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C31H39N3O4S |
|---|
| Mol. Mass. | 549.724 |
|---|
| SMILES | CCCN(c1cc2COC[C@](C)(N)Cc3cccc(CC[C@@H](NC(=O)c(c2)c1)c1ccccc1)c3)S(C)(=O)=O |
|---|
| Structure | 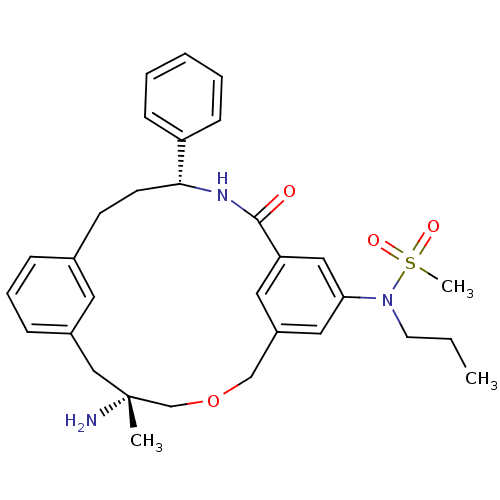
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lindsley, SR; Moore, KP; Rajapakse, HA; Selnick, HG; Young, MB; Zhu, H; Munshi, S; Kuo, L; McGaughey, GB; Colussi, D; Crouthamel, MC; Lai, MT; Pietrak, B; Price, EA; Sankaranarayanan, S; Simon, AJ; Seabrook, GR; Hazuda, DJ; Pudvah, NT; Hochman, JH; Graham, SL; Vacca, JP; Nantermet, PG Design, synthesis, and SAR of macrocyclic tertiary carbinamine BACE-1 inhibitors. Bioorg Med Chem Lett17:4057-61 (2007) [PubMed] Article
Lindsley, SR; Moore, KP; Rajapakse, HA; Selnick, HG; Young, MB; Zhu, H; Munshi, S; Kuo, L; McGaughey, GB; Colussi, D; Crouthamel, MC; Lai, MT; Pietrak, B; Price, EA; Sankaranarayanan, S; Simon, AJ; Seabrook, GR; Hazuda, DJ; Pudvah, NT; Hochman, JH; Graham, SL; Vacca, JP; Nantermet, PG Design, synthesis, and SAR of macrocyclic tertiary carbinamine BACE-1 inhibitors. Bioorg Med Chem Lett17:4057-61 (2007) [PubMed] Article