| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cyclin-dependent kinase 2 |
|---|
| Ligand | BDBM7126 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_519498 (CHEMBL951804) |
|---|
| IC50 | 224±n/a nM |
|---|
| Citation |  Mascarenhas, NM; Ghoshal, N An efficient tool for identifying inhibitors based on 3D-QSAR and docking using feature-shape pharmacophore of biologically active conformation--a case study with CDK2/cyclinA. Eur J Med Chem43:2807-18 (2008) [PubMed] Article Mascarenhas, NM; Ghoshal, N An efficient tool for identifying inhibitors based on 3D-QSAR and docking using feature-shape pharmacophore of biologically active conformation--a case study with CDK2/cyclinA. Eur J Med Chem43:2807-18 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cyclin-dependent kinase 2 |
|---|
| Name: | Cyclin-dependent kinase 2 |
|---|
| Synonyms: | CDK2 | CDK2-Kinase | CDK2_HUMAN | CDKN2 | Cell division protein kinase 2 | Cyclin-dependent kinase 2 (CDK2) | Protein cereblon/Cyclin-dependent kinase 2 | p33 protein kinase |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 33938.17 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P24941 |
|---|
| Residue: | 298 |
|---|
| Sequence: | MENFQKVEKIGEGTYGVVYKARNKLTGEVVALKKIRLDTETEGVPSTAIREISLLKELNH
PNIVKLLDVIHTENKLYLVFEFLHQDLKKFMDASALTGIPLPLIKSYLFQLLQGLAFCHS
HRVLHRDLKPQNLLINTEGAIKLADFGLARAFGVPVRTYTHEVVTLWYRAPEILLGCKYY
STAVDIWSLGCIFAEMVTRRALFPGDSEIDQLFRIFRTLGTPDEVVWPGVTSMPDYKPSF
PKWARQDFSKVVPPLDEDGRSLLSQMLHYDPNKRISAKAALAHPFFQDVTKPVPHLRL
|
|
|
|---|
| BDBM7126 |
|---|
| n/a |
|---|
| Name | BDBM7126 |
|---|
| Synonyms: | 3-Propylaminopyrazole deriv. 3 | CHEMBL112442 | CS2 | N-(5-Cyclopropyl-1H-pyrazol-3-yl)butanamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C10H15N3O |
|---|
| Mol. Mass. | 193.2456 |
|---|
| SMILES | CCCC(=O)Nc1cc([nH]n1)C1CC1 |
|---|
| Structure | 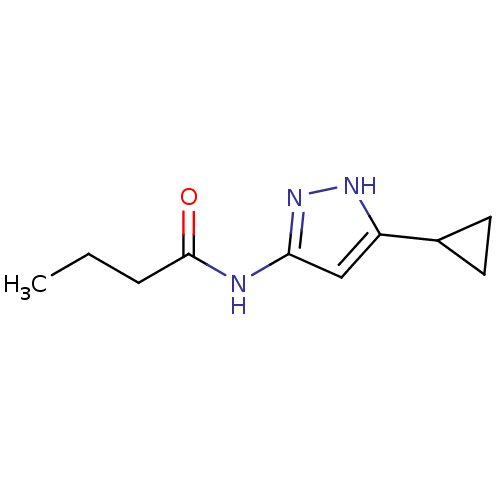
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Mascarenhas, NM; Ghoshal, N An efficient tool for identifying inhibitors based on 3D-QSAR and docking using feature-shape pharmacophore of biologically active conformation--a case study with CDK2/cyclinA. Eur J Med Chem43:2807-18 (2008) [PubMed] Article
Mascarenhas, NM; Ghoshal, N An efficient tool for identifying inhibitors based on 3D-QSAR and docking using feature-shape pharmacophore of biologically active conformation--a case study with CDK2/cyclinA. Eur J Med Chem43:2807-18 (2008) [PubMed] Article