| Reaction Details |
|---|
 | Report a problem with these data |
| Target | NH(3)-dependent NAD(+) synthetase |
|---|
| Ligand | BDBM50278299 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_564355 (CHEMBL957657) |
|---|
| IC50 | 97000±n/a nM |
|---|
| Citation |  Moro, WB; Yang, Z; Kane, TA; Brouillette, CG; Brouillette, WJ Virtual screening to identify lead inhibitors for bacterial NAD synthetase (NADs). Bioorg Med Chem Lett19:2001-5 (2009) [PubMed] Article Moro, WB; Yang, Z; Kane, TA; Brouillette, CG; Brouillette, WJ Virtual screening to identify lead inhibitors for bacterial NAD synthetase (NADs). Bioorg Med Chem Lett19:2001-5 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| NH(3)-dependent NAD(+) synthetase |
|---|
| Name: | NH(3)-dependent NAD(+) synthetase |
|---|
| Synonyms: | BAS1855 | BA_1998 | GBAA_1998 | NADE_BACAN | Nicotinamide adenine dinucleotide synthetase (NADs) | nadE |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 30092.16 |
|---|
| Organism: | Bacillus anthracis |
|---|
| Description: | Q81RP3 |
|---|
| Residue: | 272 |
|---|
| Sequence: | MTLQEQIMKALHVQPVIDPKAEIRKRVDFLKDYVKKTGAKGFVLGISGGQDSTLAGRLAQ
LAVEEIRNEGGNATFIAVRLPYKVQKDEDDAQLALQFIQADQSVAFDIASTVDAFSNQYE
NLLDESLTDFNKGNVKARIRMVTQYAIGGQKGLLVIGTDHAAEAVTGFFTKFGDGGADLL
PLTGLTKRQGRALLQELGADERLYLKMPTADLLDEKPGQADETELGITYDQLDDYLEGKT
VPADVAEKIEKRYTVSEHKRQVPASMFDDWWK
|
|
|
|---|
| BDBM50278299 |
|---|
| n/a |
|---|
| Name | BDBM50278299 |
|---|
| Synonyms: | 1-(6-(7-chloroquinazolin-4-yloxy)pyridin-3-yl)-3-(3-(cyclopentyloxy)-4-methoxyphenyl)urea | CHEMBL471230 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H24ClN5O4 |
|---|
| Mol. Mass. | 505.953 |
|---|
| SMILES | COc1ccc(NC(=O)Nc2ccc(Oc3ncnc4cc(Cl)ccc34)nc2)cc1OC1CCCC1 |
|---|
| Structure | 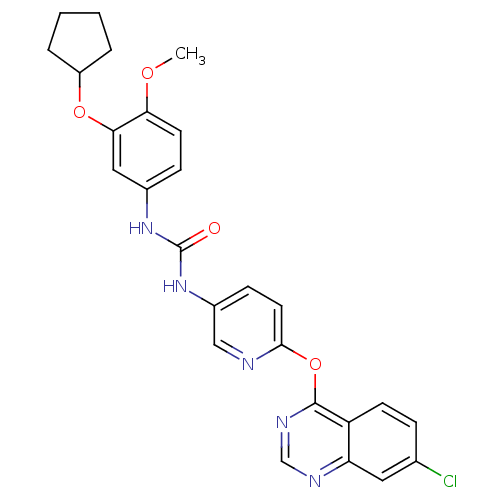
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Moro, WB; Yang, Z; Kane, TA; Brouillette, CG; Brouillette, WJ Virtual screening to identify lead inhibitors for bacterial NAD synthetase (NADs). Bioorg Med Chem Lett19:2001-5 (2009) [PubMed] Article
Moro, WB; Yang, Z; Kane, TA; Brouillette, CG; Brouillette, WJ Virtual screening to identify lead inhibitors for bacterial NAD synthetase (NADs). Bioorg Med Chem Lett19:2001-5 (2009) [PubMed] Article