| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Dipeptidyl peptidase 8 |
|---|
| Ligand | BDBM50339098 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_730296 (CHEMBL1696536) |
|---|
| Ki | 182000±n/a nM |
|---|
| Citation |  Kaczanowska, K; Wiesmüller, KH; Schaffner, AP Design, Synthesis, and in Vitro Evaluation of Novel Aminomethyl-pyridines as DPP-4 Inhibitors. ACS Med Chem Lett1:530-535 (2010) [PubMed] Article Kaczanowska, K; Wiesmüller, KH; Schaffner, AP Design, Synthesis, and in Vitro Evaluation of Novel Aminomethyl-pyridines as DPP-4 Inhibitors. ACS Med Chem Lett1:530-535 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Dipeptidyl peptidase 8 |
|---|
| Name: | Dipeptidyl peptidase 8 |
|---|
| Synonyms: | DPP8 | DPP8_HUMAN | DPRP-1 | DPRP1 | Dipeptidyl peptidase 8 (DPP-8) | Dipeptidyl peptidase 8 (DPP8) | Dipeptidyl peptidase 8/9 | Dipeptidyl peptidase IV-related protein 1 | Dipeptidyl peptidase VIII | Dipeptidyl peptidase VIII (DDP-VIII) | Prolyl dipeptidase DPP8 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 103342.62 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q6V1X1 |
|---|
| Residue: | 898 |
|---|
| Sequence: | MWKRSEQMKIKSGKCNMAAAMETEQLGVEIFETADCEENIESQDRPKLEPFYVERYSWSQ
LKKLLADTRKYHGYMMAKAPHDFMFVKRNDPDGPHSDRIYYLAMSGENRENTLFYSEIPK
TINRAAVLMLSWKPLLDLFQATLDYGMYSREEELLRERKRIGTVGIASYDYHQGSGTFLF
QAGSGIYHVKDGGPQGFTQQPLRPNLVETSCPNIRMDPKLCPADPDWIAFIHSNDIWISN
IVTREERRLTYVHNELANMEEDARSAGVATFVLQEEFDRYSGYWWCPKAETTPSGGKILR
ILYEENDESEVEIIHVTSPMLETRRADSFRYPKTGTANPKVTFKMSEIMIDAEGRIIDVI
DKELIQPFEILFEGVEYIARAGWTPEGKYAWSILLDRSQTRLQIVLISPELFIPVEDDVM
ERQRLIESVPDSVTPLIIYEETTDIWINIHDIFHVFPQSHEEEIEFIFASECKTGFRHLY
KITSILKESKYKRSSGGLPAPSDFKCPIKEEIAITSGEWEVLGRHGSNIQVDEVRRLVYF
EGTKDSPLEHHLYVVSYVNPGEVTRLTDRGYSHSCCISQHCDFFISKYSNQKNPHCVSLY
KLSSPEDDPTCKTKEFWATILDSAGPLPDYTPPEIFSFESTTGFTLYGMLYKPHDLQPGK
KYPTVLFIYGGPQVQLVNNRFKGVKYFRLNTLASLGYVVVVIDNRGSCHRGLKFEGAFKY
KMGQIEIDDQVEGLQYLASRYDFIDLDRVGIHGWSYGGYLSLMALMQRSDIFRVAIAGAP
VTLWIFYDTGYTERYMGHPDQNEQGYYLGSVAMQAEKFPSEPNRLLLLHGFLDENVHFAH
TSILLSFLVRAGKPYDLQIYPQERHSIRVPESGEHYELHLLHYLQENLGSRIAALKVI
|
|
|
|---|
| BDBM50339098 |
|---|
| n/a |
|---|
| Name | BDBM50339098 |
|---|
| Synonyms: | 5-(aminomethyl)-N-cyclopropyl-4-(2,4-difluorophenyl)-6-methylpicolinamide | CHEMBL1688419 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H17F2N3O |
|---|
| Mol. Mass. | 317.3332 |
|---|
| SMILES | Cc1nc(cc(c1CN)-c1ccc(F)cc1F)C(=O)NC1CC1 |
|---|
| Structure | 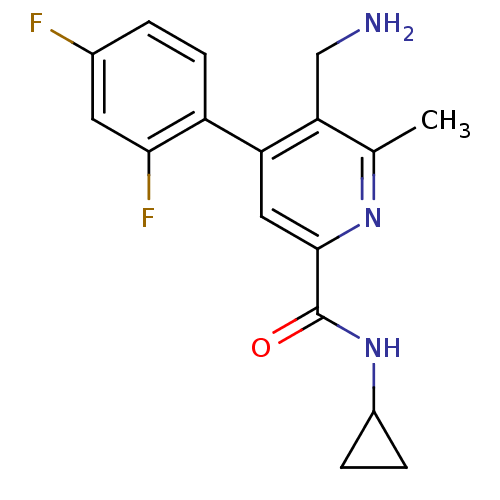
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Kaczanowska, K; Wiesmüller, KH; Schaffner, AP Design, Synthesis, and in Vitro Evaluation of Novel Aminomethyl-pyridines as DPP-4 Inhibitors. ACS Med Chem Lett1:530-535 (2010) [PubMed] Article
Kaczanowska, K; Wiesmüller, KH; Schaffner, AP Design, Synthesis, and in Vitro Evaluation of Novel Aminomethyl-pyridines as DPP-4 Inhibitors. ACS Med Chem Lett1:530-535 (2010) [PubMed] Article