

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Sodium- and chloride-dependent GABA transporter 3 | ||
| Ligand | BDBM50343258 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_745626 (CHEMBL1775996) | ||
| IC50 | >300000±n/a nM | ||
| Citation |  Hack, S; Wörlein, B; Höfner, G; Pabel, J; Wanner, KT Development of imidazole alkanoic acids as mGAT3 selective GABA uptake inhibitors. Eur J Med Chem46:1483-98 (2011) [PubMed] Article Hack, S; Wörlein, B; Höfner, G; Pabel, J; Wanner, KT Development of imidazole alkanoic acids as mGAT3 selective GABA uptake inhibitors. Eur J Med Chem46:1483-98 (2011) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Sodium- and chloride-dependent GABA transporter 3 | |||
| Name: | Sodium- and chloride-dependent GABA transporter 3 | ||
| Synonyms: | GABA transporter | GAT-3 | Gabt3 | Gabt4 | Gat-4 | Gat3 | Gat4 | S6A11_MOUSE | Slc6a11 | Sodium- and chloride-dependent GABA transporter 3 | Sodium- and chloride-dependent GABA transporter 4 | Solute carrier family 6 member 11 | ||
| Type: | PROTEIN | ||
| Mol. Mass.: | 69962.73 | ||
| Organism: | Mus musculus | ||
| Description: | ChEMBL_1460090 | ||
| Residue: | 627 | ||
| Sequence: |
| ||
| BDBM50343258 | |||
| n/a | |||
| Name | BDBM50343258 | ||
| Synonyms: | 4-((4,4-bis(3-methylthiophen-2-yl)but-3-enyl)(methyl)amino)-4,5,6,7-tetrahydrobenzo[d]isoxazol-3-ol | CHEMBL1773934 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C22H26N2O2S2 | ||
| Mol. Mass. | 414.584 | ||
| SMILES | CN(CCC=C(c1sccc1C)c1sccc1C)C1CCCc2o[nH]c(=O)c12 |(14.45,-29.58,;13.12,-28.81,;11.79,-29.58,;11.79,-31.12,;10.46,-31.89,;10.46,-33.43,;9.2,-34.32,;7.74,-33.83,;6.83,-35.07,;7.71,-36.32,;9.18,-35.86,;10.42,-36.78,;11.86,-34.07,;12.17,-35.57,;13.69,-35.75,;14.34,-34.35,;13.2,-33.31,;13.38,-31.78,;13.12,-27.27,;14.45,-26.5,;14.45,-24.97,;13.12,-24.2,;11.77,-24.97,;10.3,-24.49,;9.4,-25.74,;10.31,-26.99,;9.83,-28.46,;11.78,-26.51,)| | ||
| Structure | 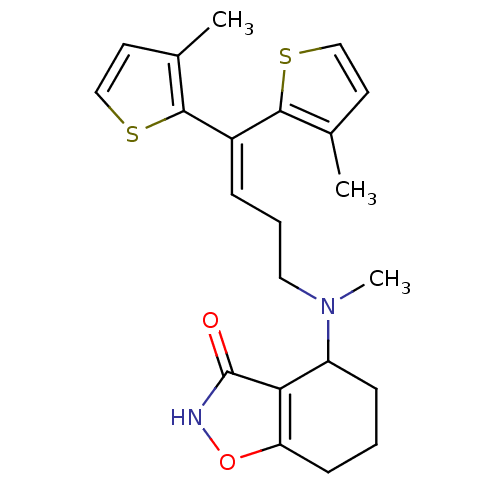
| ||