| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peptide deformylase |
|---|
| Ligand | BDBM50131332 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_154179 |
|---|
| IC50 | 30±n/a nM |
|---|
| Citation |  Davies, SJ; Ayscough, AP; Beckett, RP; Clements, JM; Doel, S; Pratt, LM; Spavold, ZM; Thomas, SW; Whittaker, M Structure--activity relationships of the peptide deformylase inhibitor BB-3497: modification of the P2' and P3' side chains. Bioorg Med Chem Lett13:2715-8 (2003) [PubMed] Davies, SJ; Ayscough, AP; Beckett, RP; Clements, JM; Doel, S; Pratt, LM; Spavold, ZM; Thomas, SW; Whittaker, M Structure--activity relationships of the peptide deformylase inhibitor BB-3497: modification of the P2' and P3' side chains. Bioorg Med Chem Lett13:2715-8 (2003) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Peptide deformylase |
|---|
| Name: | Peptide deformylase |
|---|
| Synonyms: | 3.5.1.88 | BON69_24600 | BON94_18585 | D9G11_24945 | D9G11_25760 | D9J60_20755 | FORC82_p394 | PDF | Polypeptide deformylase | SAMEA3472033_04733 | def | def_2 |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | 16901.39 |
|---|
| Organism: | Escherichia coli |
|---|
| Description: | n/a |
|---|
| Residue: | 150 |
|---|
| Sequence: | MSVLQVLHIPDERLRKVAKPVEEVNAEIQRIVDDMFETMYAEKGIGLAATQVDIHQRIIV
IDVSENRDERLVLINPELLEKSGETGIEEGCLSIPEQRALVPRAEKVKIRALDRDGKPFE
LEADGLLAICIGLRLGNGKYCTLRLFFNQV
|
|
|
|---|
| BDBM50131332 |
|---|
| n/a |
|---|
| Name | BDBM50131332 |
|---|
| Synonyms: | (R)-2-[(Formyl-hydroxy-amino)-methyl]-hexanoic acid [(S)-1-(cyclohexyl-methyl-carbamoyl)-2,2-dimethyl-propyl]-amide | CHEMBL93973 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H39N3O4 |
|---|
| Mol. Mass. | 397.5521 |
|---|
| SMILES | CCCC[C@H](CN(O)C=O)C(=O)N[C@H](C(=O)N(C)C1CCCCC1)C(C)(C)C |
|---|
| Structure | 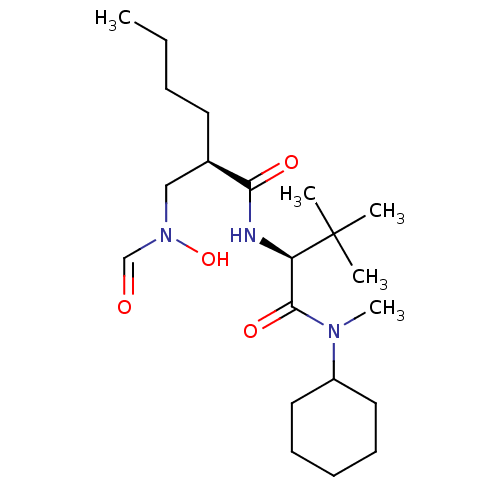
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Davies, SJ; Ayscough, AP; Beckett, RP; Clements, JM; Doel, S; Pratt, LM; Spavold, ZM; Thomas, SW; Whittaker, M Structure--activity relationships of the peptide deformylase inhibitor BB-3497: modification of the P2' and P3' side chains. Bioorg Med Chem Lett13:2715-8 (2003) [PubMed]
Davies, SJ; Ayscough, AP; Beckett, RP; Clements, JM; Doel, S; Pratt, LM; Spavold, ZM; Thomas, SW; Whittaker, M Structure--activity relationships of the peptide deformylase inhibitor BB-3497: modification of the P2' and P3' side chains. Bioorg Med Chem Lett13:2715-8 (2003) [PubMed]