

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | D(2) dopamine receptor | ||
| Ligand | BDBM50167936 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_303726 (CHEMBL829061) | ||
| Ki | 170±n/a nM | ||
| Citation |  Schlotter, K; Boeckler, F; Hübner, H; Gmeiner, P Fancy bioisosteres: metallocene-derived G-protein-coupled receptor ligands with subnanomolar binding affinity and novel selectivity profiles. J Med Chem48:3696-9 (2005) [PubMed] Article Schlotter, K; Boeckler, F; Hübner, H; Gmeiner, P Fancy bioisosteres: metallocene-derived G-protein-coupled receptor ligands with subnanomolar binding affinity and novel selectivity profiles. J Med Chem48:3696-9 (2005) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| D(2) dopamine receptor | |||
| Name: | D(2) dopamine receptor | ||
| Synonyms: | D(2) dopamine receptor | DOPAMINE D2 | DOPAMINE D2 Long | DOPAMINE D2 Short | DRD2 | DRD2_HUMAN | Dopamine D2 receptor | Dopamine D2 receptor (D2) | Dopamine D2 receptor (D2R) | Dopamine D2A | Dopamine2-like | d2 | ||
| Type: | Cell-surface receptors | ||
| Mol. Mass.: | 50647.10 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P14416 | ||
| Residue: | 443 | ||
| Sequence: |
| ||
| BDBM50167936 | |||
| n/a | |||
| Name | BDBM50167936 | ||
| Synonyms: | (2E)-3-{1-ferra-1,1'-spirobi[pentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-2,2',4,4'-tetraen-6-yl}-N-{4-[4-(2-methoxyphenyl)piperazin-1-yl]butyl}but-2-enamide | CHEMBL371165 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C29H29FeN3O2 | ||
| Mol. Mass. | 507.404 | ||
| SMILES | c1cc(OC)c(N2CCN(CC2)CCCCNC(\C=C(/C)C23C4=C5C6=C2[Fe]34562345C6C2=C3C4=C56)=O)cc1 |c:23,25,35,39,(17.57,4.15,;17.57,2.6,;16.23,1.82,;16.23,.3,;17.57,-.48,;14.9,2.6,;13.56,1.82,;13.57,.3,;12.21,-.48,;10.88,.3,;10.9,1.85,;12.23,2.61,;9.55,-.45,;8.21,.32,;6.87,-.45,;5.54,.33,;4.19,-.43,;2.86,.34,;1.53,-.43,;.19,.34,;.19,1.89,;-1.14,-.43,;-1.14,-1.57,;-2.22,-1.93,;-2.89,-1.01,;-2.22,-.09,;-3.45,-1.88,;-5.63,-3.73,;-4.55,-4.08,;-3.9,-3.16,;-4.56,-2.24,;-5.63,-2.6,;2.86,1.89,;14.9,4.14,;16.23,4.93,)| | ||
| Structure | 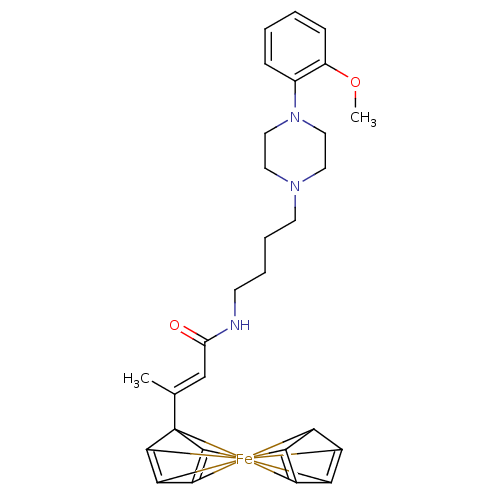
| ||