

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Muscarinic acetylcholine receptor M1 | ||
| Ligand | BDBM50175566 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_320891 (CHEMBL872390) | ||
| Ki | 6.6±n/a nM | ||
| Citation |  Naito, R; Yonetoku, Y; Okamoto, Y; Toyoshima, A; Ikeda, K; Takeuchi, M Synthesis and antimuscarinic properties of quinuclidin-3-yl 1,2,3,4-tetrahydroisoquinoline-2-carboxylate derivatives as novel muscarinic receptor antagonists. J Med Chem48:6597-606 (2005) [PubMed] Article Naito, R; Yonetoku, Y; Okamoto, Y; Toyoshima, A; Ikeda, K; Takeuchi, M Synthesis and antimuscarinic properties of quinuclidin-3-yl 1,2,3,4-tetrahydroisoquinoline-2-carboxylate derivatives as novel muscarinic receptor antagonists. J Med Chem48:6597-606 (2005) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Muscarinic acetylcholine receptor M1 | |||
| Name: | Muscarinic acetylcholine receptor M1 | ||
| Synonyms: | ACM1_RAT | Cholinergic, muscarinic M1 | Chrm-1 | Chrm1 | cholinergic receptor, muscarinic 1 | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 51390.46 | ||
| Organism: | RAT | ||
| Description: | P08482 | ||
| Residue: | 460 | ||
| Sequence: |
| ||
| BDBM50175566 | |||
| n/a | |||
| Name | BDBM50175566 | ||
| Synonyms: | 1-Cyclohexyl-3,4-dihydro-1H-isoquinoline-2-carboxylic acid (R)-(1-aza-bicyclo[2.2.2]oct-3-yl) ester | CHEMBL372983 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H32N2O2 | ||
| Mol. Mass. | 368.5124 | ||
| SMILES | O=C(O[C@H]1CN2CCC1CC2)N1CCc2ccccc2C1C1CCCCC1 |wD:3.2,(-1.26,-.59,;-1.28,.95,;.05,1.75,;1.39,1,;1.41,-.55,;2.76,-1.3,;3.43,-.15,;2,.51,;2.7,1.8,;4.06,1.05,;4.08,-.5,;-2.53,1.7,;-2.59,3.25,;-3.95,3.97,;-5.25,3.15,;-6.55,3.86,;-7.82,3.11,;-7.8,1.61,;-6.49,.89,;-5.19,1.61,;-3.83,.88,;-3.79,-.66,;-2.43,-1.39,;-2.38,-2.93,;-3.69,-3.75,;-5.06,-3.02,;-5.1,-1.48,)| | ||
| Structure | 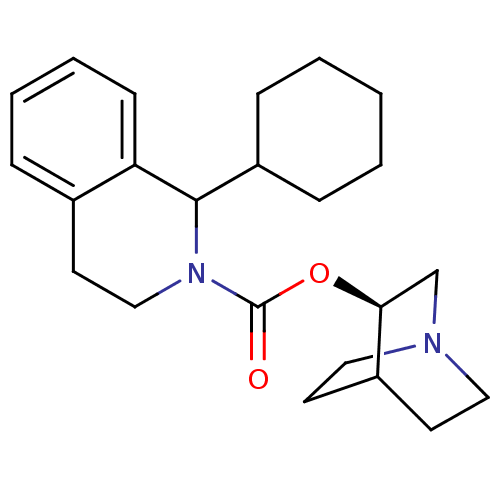
| ||