| Reaction Details |
|---|
 | Report a problem with these data |
| Target | 72 kDa type IV collagenase |
|---|
| Ligand | BDBM50225397 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_832930 (CHEMBL2066885) |
|---|
| IC50 | 4±n/a nM |
|---|
| Citation |  Hugenberg, V; Breyholz, HJ; Riemann, B; Hermann, S; Schober, O; Schäfers, M; Gangadharmath, U; Mocharla, V; Kolb, H; Walsh, J; Zhang, W; Kopka, K; Wagner, S A new class of highly potent matrix metalloproteinase inhibitors based on triazole-substituted hydroxamates: (radio)synthesis and in vitro and first in vivo evaluation. J Med Chem55:4714-27 (2012) [PubMed] Article Hugenberg, V; Breyholz, HJ; Riemann, B; Hermann, S; Schober, O; Schäfers, M; Gangadharmath, U; Mocharla, V; Kolb, H; Walsh, J; Zhang, W; Kopka, K; Wagner, S A new class of highly potent matrix metalloproteinase inhibitors based on triazole-substituted hydroxamates: (radio)synthesis and in vitro and first in vivo evaluation. J Med Chem55:4714-27 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| 72 kDa type IV collagenase |
|---|
| Name: | 72 kDa type IV collagenase |
|---|
| Synonyms: | 72 kDa gelatinase | 72 kDa type IV collagenase precursor | CLG4A | Gelatinase A | Gelatinase A (MMP-2) | MMP2 | MMP2_HUMAN | Matrix metalloproteinase-2 | Matrix metalloproteinase-2 (MMP 2) | Matrix metalloproteinase-2 (MMP2) | Matrix metalloproteinases 2 (MMP-2) | TBE-1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 73870.36 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P08253 |
|---|
| Residue: | 660 |
|---|
| Sequence: | MEALMARGALTGPLRALCLLGCLLSHAAAAPSPIIKFPGDVAPKTDKELAVQYLNTFYGC
PKESCNLFVLKDTLKKMQKFFGLPQTGDLDQNTIETMRKPRCGNPDVANYNFFPRKPKWD
KNQITYRIIGYTPDLDPETVDDAFARAFQVWSDVTPLRFSRIHDGEADIMINFGRWEHGD
GYPFDGKDGLLAHAFAPGTGVGGDSHFDDDELWTLGEGQVVRVKYGNADGEYCKFPFLFN
GKEYNSCTDTGRSDGFLWCSTTYNFEKDGKYGFCPHEALFTMGGNAEGQPCKFPFRFQGT
SYDSCTTEGRTDGYRWCGTTEDYDRDKKYGFCPETAMSTVGGNSEGAPCVFPFTFLGNKY
ESCTSAGRSDGKMWCATTANYDDDRKWGFCPDQGYSLFLVAAHEFGHAMGLEHSQDPGAL
MAPIYTYTKNFRLSQDDIKGIQELYGASPDIDLGTGPTPTLGPVTPEICKQDIVFDGIAQ
IRGEIFFFKDRFIWRTVTPRDKPMGPLLVATFWPELPEKIDAVYEAPQEEKAVFFAGNEY
WIYSASTLERGYPKPLTSLGLPPDVQRVDAAFNWSKNKKTYIFAGDKFWRYNEVKKKMDP
GFPKLIADAWNAIPDNLDAVVDLQGGGHSYFFKGAYYLKLENQSLKSVKFGSIKSDWLGC
|
|
|
|---|
| BDBM50225397 |
|---|
| n/a |
|---|
| Name | BDBM50225397 |
|---|
| Synonyms: | (R)-2-(N-benzyl-4-(2-fluoroethoxy)phenylsulfonamido)-N-hydroxy-3-methylbutanamide | CHEMBL429800 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H25FN2O5S |
|---|
| Mol. Mass. | 424.486 |
|---|
| SMILES | CC(C)[C@@H](N(Cc1ccccc1)S(=O)(=O)c1ccc(OCCF)cc1)C(=O)NO |
|---|
| Structure | 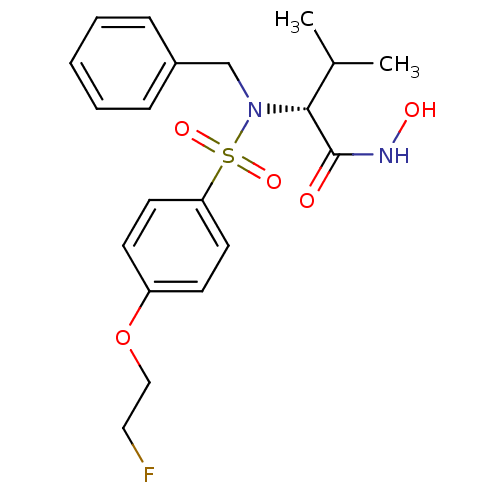
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Hugenberg, V; Breyholz, HJ; Riemann, B; Hermann, S; Schober, O; Schäfers, M; Gangadharmath, U; Mocharla, V; Kolb, H; Walsh, J; Zhang, W; Kopka, K; Wagner, S A new class of highly potent matrix metalloproteinase inhibitors based on triazole-substituted hydroxamates: (radio)synthesis and in vitro and first in vivo evaluation. J Med Chem55:4714-27 (2012) [PubMed] Article
Hugenberg, V; Breyholz, HJ; Riemann, B; Hermann, S; Schober, O; Schäfers, M; Gangadharmath, U; Mocharla, V; Kolb, H; Walsh, J; Zhang, W; Kopka, K; Wagner, S A new class of highly potent matrix metalloproteinase inhibitors based on triazole-substituted hydroxamates: (radio)synthesis and in vitro and first in vivo evaluation. J Med Chem55:4714-27 (2012) [PubMed] Article