

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Voltage-dependent L-type calcium channel subunit alpha-1C | ||
| Ligand | BDBM50370682 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1478910 (CHEMBL3430382) | ||
| IC50 | 6310±n/a nM | ||
| Citation |  Mirams, GR; Davies, MR; Brough, SJ; Bridgland-Taylor, MH; Cui, Y; Gavaghan, DJ; Abi-Gerges, N Prediction of Thorough QT study results using action potential simulations based on ion channel screens. J Pharmacol Toxicol Methods70:246-54 (2014) [PubMed] Article Mirams, GR; Davies, MR; Brough, SJ; Bridgland-Taylor, MH; Cui, Y; Gavaghan, DJ; Abi-Gerges, N Prediction of Thorough QT study results using action potential simulations based on ion channel screens. J Pharmacol Toxicol Methods70:246-54 (2014) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Voltage-dependent L-type calcium channel subunit alpha-1C | |||
| Name: | Voltage-dependent L-type calcium channel subunit alpha-1C | ||
| Synonyms: | CAC1C_HUMAN | CACH2 | CACN2 | CACNA1C | CACNL1A1 | CCHL1A1 | Calcium channel (Type L) | Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle | L-type calcium channel alpha-1c/beta-2/alpha2delta-1 | Voltage-dependent L-type calcium channel subunit alpha-1C | Voltage-gated L-type calcium channel | Voltage-gated L-type calcium channel alpha-1C subunit | Voltage-gated calcium channel | Voltage-gated calcium channel subunit alpha Cav1.2 | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 248979.79 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Calcium channel (Type L) 0 HUMAN::Q13936 | ||
| Residue: | 2221 | ||
| Sequence: |
| ||
| BDBM50370682 | |||
| n/a | |||
| Name | BDBM50370682 | ||
| Synonyms: | SOLIFENACIN | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H26N2O2 | ||
| Mol. Mass. | 362.4647 | ||
| SMILES | [H]C12CCN(CC1)C[C@@H]2OC(=O)N1CCc2ccccc2[C@@H]1c1ccccc1 |wD:8.10,21.25,TLB:9:8:2.3:6.5,(23.54,-9.18,;23.57,-7.64,;22.04,-8.3,;21.84,-6.92,;23.31,-6.28,;23.38,-4.63,;23.82,-5.74,;24.65,-6.88,;24.94,-8.28,;24.93,-9.81,;23.59,-10.56,;22.26,-9.79,;23.59,-12.1,;24.92,-12.9,;24.88,-14.45,;23.53,-15.19,;23.52,-16.72,;22.17,-17.47,;20.85,-16.68,;20.87,-15.14,;22.21,-14.39,;22.23,-12.84,;20.91,-12.06,;19.57,-12.81,;18.25,-12.02,;18.28,-10.47,;19.62,-9.73,;20.94,-10.52,)| | ||
| Structure | 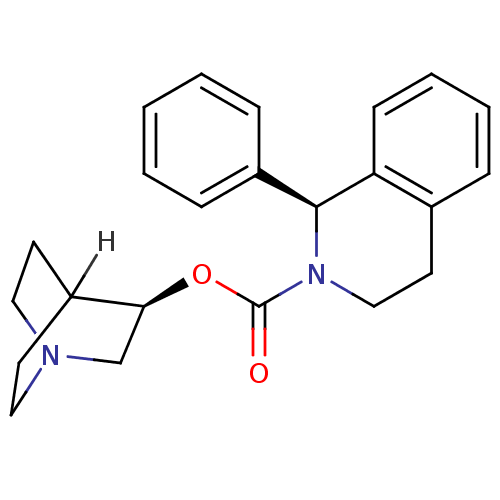
| ||