| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Glutamate receptor ionotropic, NMDA 1 |
|---|
| Ligand | BDBM85822 |
|---|
| Substrate/Competitor | n/a |
|---|
| Ki | >10000±n/a nM |
|---|
| Comments | PDSP_3686 |
|---|
| Citation |  Johansen, TN; Stensbøl, TB; Nielsen, B; Vogensen, SB; Frydenvang, K; Sløk, FA; Bräüner-Osborne, H; Madsen, U; Krogsgaard-Larsen, P Resolution, configurational assignment, and enantiopharmacology at glutamate receptors of 2-amino-3-(3-carboxy-5-methyl-4-isoxazolyl)propionic acid (ACPA) and demethyl-ACPA. Chirality13:523-32 (2001) [PubMed] Article Johansen, TN; Stensbøl, TB; Nielsen, B; Vogensen, SB; Frydenvang, K; Sløk, FA; Bräüner-Osborne, H; Madsen, U; Krogsgaard-Larsen, P Resolution, configurational assignment, and enantiopharmacology at glutamate receptors of 2-amino-3-(3-carboxy-5-methyl-4-isoxazolyl)propionic acid (ACPA) and demethyl-ACPA. Chirality13:523-32 (2001) [PubMed] Article |
|---|
| More Info.: | Get all data from this article |
|---|
| |
| Glutamate receptor ionotropic, NMDA 1 |
|---|
| Name: | Glutamate receptor ionotropic, NMDA 1 |
|---|
| Synonyms: | Glutamate (NMDA) receptor subunit zeta 1 | Glutamate [NMDA] receptor subunit zeta-1 | Glutamate-NMDA-Channel | Glutamate-NMDA-MK801 | Glutamate-NMDA-Polyamine | Grin1 | NMDA | NMDZ1_RAT | Nmdar1 | phencyclidine |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 105533.40 |
|---|
| Organism: | RAT |
|---|
| Description: | P35439 |
|---|
| Residue: | 938 |
|---|
| Sequence: | MSTMHLLTFALLFSCSFARAACDPKIVNIGAVLSTRKHEQMFREAVNQANKRHGSWKIQL
NATSVTHKPNAIQMALSVCEDLISSQVYAILVSHPPTPNDHFTPTPVSYTAGFYRIPVLG
LTTRMSIYSDKSIHLSFLRTVPPYSHQSSVWFEMMRVYNWNHIILLVSDDHEGRAAQKRL
ETLLEERESKAEKVLQFDPGTKNVTALLMEARELEARVIILSASEDDAATVYRAAAMLNM
TGSGYVWLVGEREISGNALRYAPDGIIGLQLINGKNESAHISDAVGVVAQAVHELLEKEN
ITDPPRGCVGNTNIWKTGPLFKRVLMSSKYADGVTGRVEFNEDGDRKFANYSIMNLQNRK
LVQVGIYNGTHVIPNDRKIIWPGGETEKPRGYQMSTRLKIVTIHQEPFVYVKPTMSDGTC
KEEFTVNGDPVKKVICTGPNDTSPGSPRHTVPQCCYGFCIDLLIKLARTMNFTYEVHLVA
DGKFGTQERVNNSNKKEWNGMMGELLSGQADMIVAPLTINNERAQYIEFSKPFKYQGLTI
LVKKEIPRSTLDSFMQPFQSTLWLLVGLSVHVVAVMLYLLDRFSPFGRFKVNSEEEEEDA
LTLSSAMWFSWGVLLNSGIGEGAPRSFSARILGMVWAGFAMIIVASYTANLAAFLVLDRP
EERITGINDPRLRNPSDKFIYATVKQSSVDIYFRRQVELSTMYRHMEKHNYESAAEAIQA
VRDNKLHAFIWDSAVLEFEASQKCDLVTTGELFFRSGFGIGMRKDSPWKQNVSLSILKSH
ENGFMEDLDKTWVRYQECDSRSNAPATLTFENMAGVFMLVAGGIVAGIFLIFIEIAYKRH
KDARRKQMQLAFAAVNVWRKNLQDRKSGRAEPDPKKKATFRAITSTLASSFKRRRSSKDT
STGGGRGALQNQKDTVLPRRAIEREEGQLQLCSRHRES
|
|
|
|---|
| BDBM85822 |
|---|
| n/a |
|---|
| Name | BDBM85822 |
|---|
| Synonyms: | Demethyl-AMPA | OFQ/N | OFQ/N (1-11) | OFQ/N (1-11),[Tyr10] |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C61H100N22O15 |
|---|
| Mol. Mass. | 1381.5851 |
|---|
| SMILES | [#6]-[#6@@H](-[#8])-[#6@H](-[#7]-[#6](=O)-[#6@H](-[#6]-c1ccccc1)-[#7]-[#6](=O)-[#6]-[#7]-[#6](=O)-[#6]-[#7]-[#6](=O)-[#6@@H](-[#7])-[#6]-c1ccccc1)-[#6](=O)-[#7]-[#6]-[#6](=O)-[#7]-[#6@@H](-[#6])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]\[#7]=[#6](\[#7])-[#7])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]-[#6]-[#7])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#8])-[#6](=O)-[#7]-[#6@@H](-[#6])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]\[#7]=[#6](\[#7])-[#7])-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]-[#6]-[#7])-[#6](-[#7])=O |
|---|
| Structure | 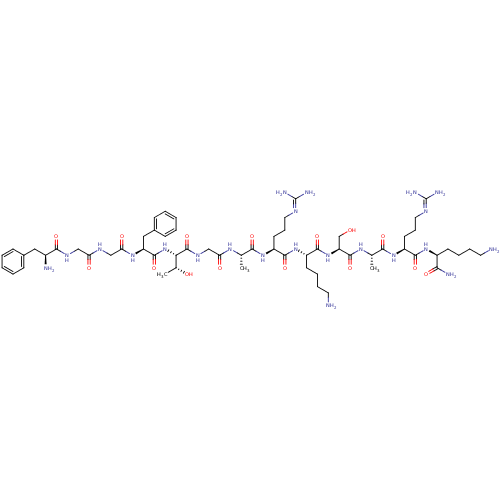
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Johansen, TN; Stensbøl, TB; Nielsen, B; Vogensen, SB; Frydenvang, K; Sløk, FA; Bräüner-Osborne, H; Madsen, U; Krogsgaard-Larsen, P Resolution, configurational assignment, and enantiopharmacology at glutamate receptors of 2-amino-3-(3-carboxy-5-methyl-4-isoxazolyl)propionic acid (ACPA) and demethyl-ACPA. Chirality13:523-32 (2001) [PubMed] Article
Johansen, TN; Stensbøl, TB; Nielsen, B; Vogensen, SB; Frydenvang, K; Sløk, FA; Bräüner-Osborne, H; Madsen, U; Krogsgaard-Larsen, P Resolution, configurational assignment, and enantiopharmacology at glutamate receptors of 2-amino-3-(3-carboxy-5-methyl-4-isoxazolyl)propionic acid (ACPA) and demethyl-ACPA. Chirality13:523-32 (2001) [PubMed] Article