| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Enoyl-[acyl-carrier-protein] reductase [NADH] |
|---|
| Ligand | BDBM8726 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Inhibition Kinetics Assay |
|---|
| pH | 8±0 |
|---|
| Temperature | 298.15±0 K |
|---|
| Ki | 32±0 nM |
|---|
| Citation |  Neckles, C; Eltschkner, S; Cummings, JE; Hirschbeck, M; Daryaee, F; Bommineni, GR; Zhang, Z; Spagnuolo, L; Yu, W; Davoodi, S; Slayden, RA; Kisker, C; Tonge, PJ Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase. Biochemistry56:1865-1878 (2017) [PubMed] Article Neckles, C; Eltschkner, S; Cummings, JE; Hirschbeck, M; Daryaee, F; Bommineni, GR; Zhang, Z; Spagnuolo, L; Yu, W; Davoodi, S; Slayden, RA; Kisker, C; Tonge, PJ Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase. Biochemistry56:1865-1878 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Enoyl-[acyl-carrier-protein] reductase [NADH] |
|---|
| Name: | Enoyl-[acyl-carrier-protein] reductase [NADH] |
|---|
| Synonyms: | Enoyl-ACP reductase (FabI1) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 27824.56 |
|---|
| Organism: | Burkholderia pseudomallei |
|---|
| Description: | Q3JG55 |
|---|
| Residue: | 253 |
|---|
| Sequence: | MRLQHKRGLIIGIANENSIAFGCARVMREQGAELALTYLNEKAEPYVRPLAQRLDSRLVV
PCDVREPGRLEDVFARIAQEWGQLDFVLHSIAYAPKEDLHRRVTDCSQAGFAMAMDVSCH
SFIRVARLAEPLMTNGGCLLTVTFYGAERAVEDYNLMGPVKAALEGSVRYLAAELGPRRI
RVHALSPGPLKTRAASGIDRFDALLERVRERTPGHRLVDIDDVGHVAAFLASDDAAALTG
NVEYIDGGYHVVG
|
|
|
|---|
| BDBM8726 |
|---|
| n/a |
|---|
| Name | BDBM8726 |
|---|
| Synonyms: | 5-chloro-2-(2,4-dichlorophenoxy)phenol | CHEMBL849 | TCL | Triclosan | US20230414581, Compound 35 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C12H7Cl3O2 |
|---|
| Mol. Mass. | 289.542 |
|---|
| SMILES | Oc1cc(Cl)ccc1Oc1ccc(Cl)cc1Cl |
|---|
| Structure | 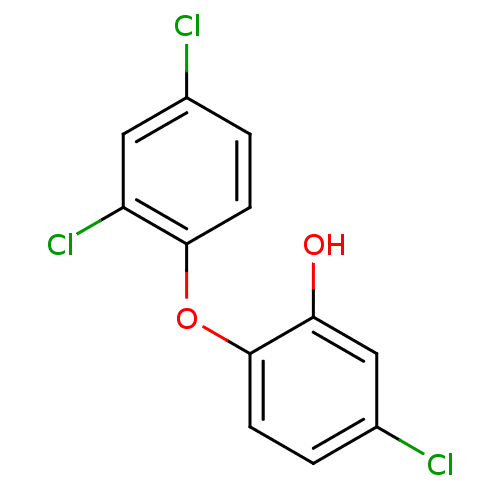
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Neckles, C; Eltschkner, S; Cummings, JE; Hirschbeck, M; Daryaee, F; Bommineni, GR; Zhang, Z; Spagnuolo, L; Yu, W; Davoodi, S; Slayden, RA; Kisker, C; Tonge, PJ Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase. Biochemistry56:1865-1878 (2017) [PubMed] Article
Neckles, C; Eltschkner, S; Cummings, JE; Hirschbeck, M; Daryaee, F; Bommineni, GR; Zhang, Z; Spagnuolo, L; Yu, W; Davoodi, S; Slayden, RA; Kisker, C; Tonge, PJ Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase. Biochemistry56:1865-1878 (2017) [PubMed] Article