| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Isocitrate dehydrogenase [NADP] cytoplasmic [R132H] |
|---|
| Ligand | BDBM287893 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | R132H IDH1 Enzymatic Assay |
|---|
| IC50 | 18.0±n/a nM |
|---|
| Citation |  Fischer, C; Bogen, SL; Childers, ML; Fradera Llinas, FX; Ellis, JM; Esposite, S; Hoffman, DM; Huang, C; Kattar, SD; Kim, AJ; Lampe, JW; Machacek, MR; McMasters, DR; Parker, Jr., DL; Reutershan, MH; Sciammetta, N; Shao, PP; Sloman, DL; Sun, W; Ujjainwalla, F; Wu, Z; Yu, Y; Gibeau, CR Tricyclic compounds as inhibitors of mutant IDH enzymes US Patent US10508108 Publication Date 12/17/2019 Fischer, C; Bogen, SL; Childers, ML; Fradera Llinas, FX; Ellis, JM; Esposite, S; Hoffman, DM; Huang, C; Kattar, SD; Kim, AJ; Lampe, JW; Machacek, MR; McMasters, DR; Parker, Jr., DL; Reutershan, MH; Sciammetta, N; Shao, PP; Sloman, DL; Sun, W; Ujjainwalla, F; Wu, Z; Yu, Y; Gibeau, CR Tricyclic compounds as inhibitors of mutant IDH enzymes US Patent US10508108 Publication Date 12/17/2019 |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Isocitrate dehydrogenase [NADP] cytoplasmic [R132H] |
|---|
| Name: | Isocitrate dehydrogenase [NADP] cytoplasmic [R132H] |
|---|
| Synonyms: | Cytosolic NADP-isocitrate dehydrogenase (IDH1)(R132H) | IDH1 | IDH1 R132H | IDH1(R132H) | IDHC_HUMAN | Isocitrate dehydrogenase (IDH1)(R132H) | Isocitrate dehydrogenase 1 mutant (R132H) | Isocitrate dehydrogenase [NADP] cytoplasmic (IDH)(R132H) | Isocitrate dehydrogenase [NADP] cytoplasmic (IDH1)(R132H) | Isocitrate dehydrogenase [NADP] cytoplasmic (R132H) | PICD |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 46641.74 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Human IDH1 R132H (SEQ ID No. 2 in patent). First three are removed. Google patent parsed wrong. |
|---|
| Residue: | 414 |
|---|
| Sequence: | MSKKISGGSVVEMQGDEMTRIIWELIKEKLIFPYVELDLHSYDLGIENRDATNDQVTKDA
AEAIKKHNVGVKCATITPDEKRVEEFKLKQMWKSPNGTIRNILGGTVFREAIICKNIPRL
VSGWVKPIIIGHHAYGDQYRATDFVVPGPGKVEITYTPSDGTQKVTYLVHNFEEGGGVAM
GMYNQDKSIEDFAHSSFQMALSKGWPLYLSTKNTILKKYDGRFKDIFQEIYDKQYKSQFE
AQKIWYEHRLIDDMVAQAMKSEGGFIWACKNYDGDVQSDSVAQGYGSLGMMTSVLVCPDG
KTVEAEAAHGTVTRHYRMYQKGQETSTNPIASIFAWTRGLAHRAKLDNNKELAFFANALE
EVSIETIEAGFMTKDLAACIKGLPNVQRSDYLNTFEFMDKLGENLKIKLAQAKL
|
|
|
|---|
| BDBM287893 |
|---|
| n/a |
|---|
| Name | BDBM287893 |
|---|
| Synonyms: | 2-Ethoxy-11-(6,7,8,9-tetrahydro-5H-benzo[7]annulen-7-ylcarbonyl)-10,11-dihydro-5H-dipyrido[2,3-b:2′,3′-e][1,4]diazepine | US10086000, Example 110 | US10508108, Example 110 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C25H26N4O2 |
|---|
| Mol. Mass. | 414.4995 |
|---|
| SMILES | CCOc1ccc2Nc3ncccc3CN(C(=O)C3CCc4ccccc4CC3)c2n1 |
|---|
| Structure | 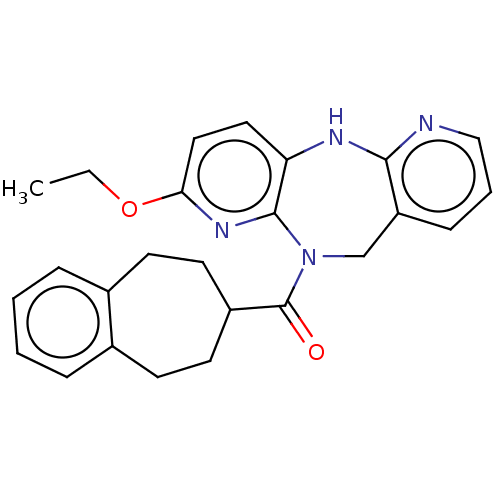
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Fischer, C; Bogen, SL; Childers, ML; Fradera Llinas, FX; Ellis, JM; Esposite, S; Hoffman, DM; Huang, C; Kattar, SD; Kim, AJ; Lampe, JW; Machacek, MR; McMasters, DR; Parker, Jr., DL; Reutershan, MH; Sciammetta, N; Shao, PP; Sloman, DL; Sun, W; Ujjainwalla, F; Wu, Z; Yu, Y; Gibeau, CR Tricyclic compounds as inhibitors of mutant IDH enzymes US Patent US10508108 Publication Date 12/17/2019
Fischer, C; Bogen, SL; Childers, ML; Fradera Llinas, FX; Ellis, JM; Esposite, S; Hoffman, DM; Huang, C; Kattar, SD; Kim, AJ; Lampe, JW; Machacek, MR; McMasters, DR; Parker, Jr., DL; Reutershan, MH; Sciammetta, N; Shao, PP; Sloman, DL; Sun, W; Ujjainwalla, F; Wu, Z; Yu, Y; Gibeau, CR Tricyclic compounds as inhibitors of mutant IDH enzymes US Patent US10508108 Publication Date 12/17/2019