| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Receptor-interacting serine/threonine-protein kinase 1 |
|---|
| Ligand | BDBM534562 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | RIPK1 HTRF Binding Assay |
|---|
| IC50 | 0.50±n/a nM |
|---|
| Citation |  Guanglin, L; Jie, C; Diane, DC; B., FD; Junqing, G; C., HA; Xirui, H; E., MM; Reiser, PM; Jianliang, S; H., SS; Lee, VB; Yong-Jin, W; Zili, X; G., YM TRIAZOLOPYRIDINYL COMPOUNDS AS KINASE INHIBITORS WIPOWO2022086828 Publication Date 4/28/2022 Guanglin, L; Jie, C; Diane, DC; B., FD; Junqing, G; C., HA; Xirui, H; E., MM; Reiser, PM; Jianliang, S; H., SS; Lee, VB; Yong-Jin, W; Zili, X; G., YM TRIAZOLOPYRIDINYL COMPOUNDS AS KINASE INHIBITORS WIPOWO2022086828 Publication Date 4/28/2022 |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Receptor-interacting serine/threonine-protein kinase 1 |
|---|
| Name: | Receptor-interacting serine/threonine-protein kinase 1 |
|---|
| Synonyms: | Cell death protein RIP | RIP | RIP-1 | RIP1 | RIPK1 | RIPK1_HUMAN | Receptor-interacting protein 1 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 75926.99 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q13546 |
|---|
| Residue: | 671 |
|---|
| Sequence: | MQPDMSLNVIKMKSSDFLESAELDSGGFGKVSLCFHRTQGLMIMKTVYKGPNCIEHNEAL
LEEAKMMNRLRHSRVVKLLGVIIEEGKYSLVMEYMEKGNLMHVLKAEMSTPLSVKGRIIL
EIIEGMCYLHGKGVIHKDLKPENILVDNDFHIKIADLGLASFKMWSKLNNEEHNELREVD
GTAKKNGGTLYYMAPEHLNDVNAKPTEKSDVYSFAVVLWAIFANKEPYENAICEQQLIMC
IKSGNRPDVDDITEYCPREIISLMKLCWEANPEARPTFPGIEEKFRPFYLSQLEESVEED
VKSLKKEYSNENAVVKRMQSLQLDCVAVPSSRSNSATEQPGSLHSSQGLGMGPVEESWFA
PSLEHPQEENEPSLQSKLQDEANYHLYGSRMDRQTKQQPRQNVAYNREEERRRRVSHDPF
AQQRPYENFQNTEGKGTAYSSAASHGNAVHQPSGLTSQPQVLYQNNGLYSSHGFGTRPLD
PGTAGPRVWYRPIPSHMPSLHNIPVPETNYLGNTPTMPFSSLPPTDESIKYTIYNSTGIQ
IGAYNYMEIGGTSSSLLDSTNTNFKEEPAAKYQAIFDNTTSLTDKHLDPIRENLGKHWKN
CARKLGFTQSQIDEIDHDYERDGLKEKVYQMLQKWVMREGIKGATVGKLAQALHQCSRID
LLSSLIYVSQN
|
|
|
|---|
| BDBM534562 |
|---|
| n/a |
|---|
| Name | BDBM534562 |
|---|
| Synonyms: | 3-(2-amino-[1,2,4]triazolo[1,5-α]pyri din-7-yl)-6-chl oro-2 -fluoro-N-((2R,3S)-2-fluoro-3-(4-fluorophenyl)-3-hydroxypropyl)benzamide | WO2022086828, Example 298 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H17ClF3N5O2 |
|---|
| Mol. Mass. | 475.851 |
|---|
| SMILES | Nc1nc2cc(ccn2n1)-c1ccc(Cl)c(C(=O)NC[C@@H](F)[C@@H](O)c2ccc(F)cc2)c1F |r| |
|---|
| Structure | 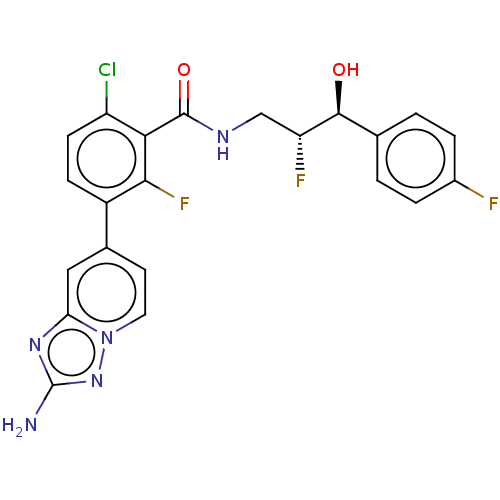
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Guanglin, L; Jie, C; Diane, DC; B., FD; Junqing, G; C., HA; Xirui, H; E., MM; Reiser, PM; Jianliang, S; H., SS; Lee, VB; Yong-Jin, W; Zili, X; G., YM TRIAZOLOPYRIDINYL COMPOUNDS AS KINASE INHIBITORS WIPOWO2022086828 Publication Date 4/28/2022
Guanglin, L; Jie, C; Diane, DC; B., FD; Junqing, G; C., HA; Xirui, H; E., MM; Reiser, PM; Jianliang, S; H., SS; Lee, VB; Yong-Jin, W; Zili, X; G., YM TRIAZOLOPYRIDINYL COMPOUNDS AS KINASE INHIBITORS WIPOWO2022086828 Publication Date 4/28/2022