| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Ubiquitin carboxyl-terminal hydrolase 2 |
|---|
| Ligand | BDBM50242207 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Fluorometric Ub-AMC Hydrolysis Assay |
|---|
| pH | 7.5±n/a |
|---|
| Temperature | 298.15±n/a K |
|---|
| IC50 | 67800±n/a nM |
|---|
| Comments | extracted |
|---|
| Citation |  Ott, CA; Baljinnyam, B; Zakharov, AV; Jadhav, A; Simeonov, A; Zhuang, Z Cell Lysate-Based AlphaLISA Deubiquitinase Assay Platform for Identification of Small Molecule Inhibitors. ACS Chem Biol12:2399-2407 (2017) [PubMed] Article Ott, CA; Baljinnyam, B; Zakharov, AV; Jadhav, A; Simeonov, A; Zhuang, Z Cell Lysate-Based AlphaLISA Deubiquitinase Assay Platform for Identification of Small Molecule Inhibitors. ACS Chem Biol12:2399-2407 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Ubiquitin carboxyl-terminal hydrolase 2 |
|---|
| Name: | Ubiquitin carboxyl-terminal hydrolase 2 |
|---|
| Synonyms: | 41 kDa ubiquitin-specific protease | Deubiquitinating enzyme 2 | UBP2_HUMAN | UBP41 | USP2 | Ubiquitin thioesterase 2 | Ubiquitin-specific-processing protease 2 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 68093.61 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_103660 |
|---|
| Residue: | 605 |
|---|
| Sequence: | MSQLSSTLKRYTESARYTDAHYAKSGYGAYTPSSYGANLAASLLEKEKLGFKPVPTSSFL
TRPRTYGPSSLLDYDRGRPLLRPDITGGGKRAESQTRGTERPLGSGLSGGSGFPYGVTNN
CLSYLPINAYDQGVTLTQKLDSQSDLARDFSSLRTSDSYRIDPRNLGRSPMLARTRKELC
TLQGLYQTASCPEYLVDYLENYGRKGSASQVPSQAPPSRVPEIISPTYRPIGRYTLWETG
KGQAPGPSRSSSPGRDGMNSKSAQGLAGLRNLGNTCFMNSILQCLSNTRELRDYCLQRLY
MRDLHHGSNAHTALVEEFAKLIQTIWTSSPNDVVSPSEFKTQIQRYAPRFVGYNQQDAQE
FLRFLLDGLHNEVNRVTLRPKSNPENLDHLPDDEKGRQMWRKYLEREDSRIGDLFVGQLK
SSLTCTDCGYCSTVFDPFWDLSLPIAKRGYPEVTLMDCMRLFTKEDVLDGDEKPTCCRCR
GRKRCIKKFSIQRFPKILVLHLKRFSESRIRTSKLTTFVNFPLRDLDLREFASENTNHAV
YNLYAVSNHSGTTMGGHYTAYCRSPGTGEWHTFNDSSVTPMSSSQVRTSDAYLLFYELAS
PPSRM
|
|
|
|---|
| BDBM50242207 |
|---|
| n/a |
|---|
| Name | BDBM50242207 |
|---|
| Synonyms: | CHEMBL455364 | Mangiferin |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H18O12 |
|---|
| Mol. Mass. | 438.339 |
|---|
| SMILES | OC[C@H]1O[C@@H](Oc2c(O)cc3oc4cc(O)c(O)cc4c(=O)c3c2O)[C@H](O)[C@@H](O)[C@@H]1O |r| |
|---|
| Structure | 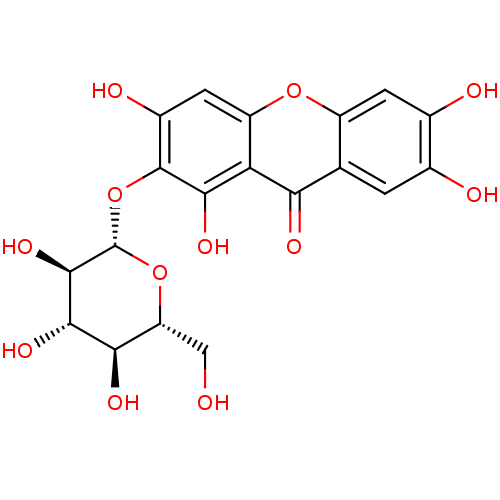
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ott, CA; Baljinnyam, B; Zakharov, AV; Jadhav, A; Simeonov, A; Zhuang, Z Cell Lysate-Based AlphaLISA Deubiquitinase Assay Platform for Identification of Small Molecule Inhibitors. ACS Chem Biol12:2399-2407 (2017) [PubMed] Article
Ott, CA; Baljinnyam, B; Zakharov, AV; Jadhav, A; Simeonov, A; Zhuang, Z Cell Lysate-Based AlphaLISA Deubiquitinase Assay Platform for Identification of Small Molecule Inhibitors. ACS Chem Biol12:2399-2407 (2017) [PubMed] Article