| Reaction Details |
|---|
 | Report a problem with these data |
| Target | ATP-sensitive inward rectifier potassium channel 1 |
|---|
| Ligand | BDBM73215 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | High throughput discovery of novel modulators of ROMK K+ channel activity: Analog Library Testing |
|---|
| IC50 | 272.6±n/a nM |
|---|
| Citation |  PubChem, PC High throughput discovery of novel modulators of ROMK K+ channel activity: Analog Library Testing PubChem Bioassay(2010)[AID] PubChem, PC High throughput discovery of novel modulators of ROMK K+ channel activity: Analog Library Testing PubChem Bioassay(2010)[AID] |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| ATP-sensitive inward rectifier potassium channel 1 |
|---|
| Name: | ATP-sensitive inward rectifier potassium channel 1 |
|---|
| Synonyms: | ATP-regulated potassium channel ROM-K | ATP-regulated potassium channel ROMK | ATP-sensitive inward rectifier potassium channel 1 | Egl nine homolog 1 | KCNJ1 | KCNJ1_HUMAN | Potassium channel (ATP modulatory) | Potassium inwardly-rectifying channel, subfamily J, member 1 | ROMK1 | Renal Outer Medullary Potassium (ROMK1) | The Renal Outer Medullary Potassium (ROMK) |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 44809.08 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | gi_223460826 |
|---|
| Residue: | 391 |
|---|
| Sequence: | MNASSRNVFDTLIRVLTESMFKHLRKWVVTRFFGHSRQRARLVSKDGRCNIEFGNVEAQS
RFIFFVDIWTTVLDLKWRYKMTIFITAFLGSWFFFGLLWYAVAYIHKDLPEFHPSANHTP
CVENINGLTSAFLFSLETQVTIGYGFRCVTEQCATAIFLLIFQSILGVIINSFMCGAILA
KISRPKKRAKTITFSKNAVISKRGGKLCLLIRVANLRKSLLIGSHIYGKLLKTTVTPEGE
TIILDQININFVVDAGNENLFFISPLTIYHVIDHNSPFFHMAAETLLQQDFELVVFLDGT
VESTSATCQVRTSYVPEEVLWGYRFAPIVSKTKEGKYRVDFHNFSKTVEVETPHCAMCLY
NEKDVRARMKRGYDNPNFILSEVNETDDTKM
|
|
|
|---|
| BDBM73215 |
|---|
| n/a |
|---|
| Name | BDBM73215 |
|---|
| Synonyms: | 7,13-bis(4-nitrobenzyl)-1,4,10-trioxa-7,13-diazacyclopentadecane;2,2,2-trifluoroacetic acid | 7,13-bis[(4-nitrophenyl)methyl]-1,4,10-trioxa-7,13-diazacyclopentadecane;2,2,2-trifluoroacetic acid | 7,13-bis[(4-nitrophenyl)methyl]-1,4,10-trioxa-7,13-diazacyclopentadecane;2,2,2-tris(fluoranyl)ethanoic acid | CHEMBL1401670 | VU0277590-7 | cid_44176372 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H32N4O7 |
|---|
| Mol. Mass. | 488.5335 |
|---|
| SMILES | [O-][N+](=O)c1ccc(CN2CCOCCOCCN(Cc3ccc(cc3)[N+]([O-])=O)CCOCC2)cc1 |
|---|
| Structure | 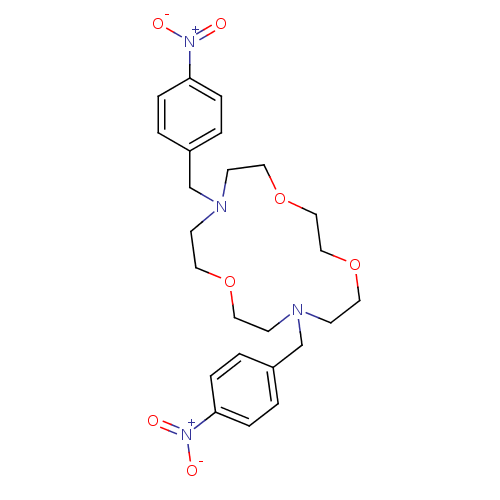
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC High throughput discovery of novel modulators of ROMK K+ channel activity: Analog Library Testing PubChem Bioassay(2010)[AID]
PubChem, PC High throughput discovery of novel modulators of ROMK K+ channel activity: Analog Library Testing PubChem Bioassay(2010)[AID]