| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Target of rapamycin complex 2 subunit MAPKAP1 |
|---|
| Ligand | BDBM50505422 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1822731 (CHEMBL4322495) |
|---|
| IC50 | 409±n/a nM |
|---|
| Citation |  Borsari, C; Rageot, D; Dall'Asen, A; Bohnacker, T; Melone, A; Sele, AM; Jackson, E; Langlois, JB; Beaufils, F; Hebeisen, P; Fabbro, D; Hillmann, P; Wymann, MP A Conformational Restriction Strategy for the Identification of a Highly Selective Pyrimido-pyrrolo-oxazine mTOR Inhibitor. J Med Chem62:8609-8630 (2019) [PubMed] Article Borsari, C; Rageot, D; Dall'Asen, A; Bohnacker, T; Melone, A; Sele, AM; Jackson, E; Langlois, JB; Beaufils, F; Hebeisen, P; Fabbro, D; Hillmann, P; Wymann, MP A Conformational Restriction Strategy for the Identification of a Highly Selective Pyrimido-pyrrolo-oxazine mTOR Inhibitor. J Med Chem62:8609-8630 (2019) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Target of rapamycin complex 2 subunit MAPKAP1 |
|---|
| Name: | Target of rapamycin complex 2 subunit MAPKAP1 |
|---|
| Synonyms: | MAPKAP1 | MIP1 | Mitogen-activated protein kinase 2-associated protein 1 | SAPK-interacting protein 1 | SIN1 | SIN1_HUMAN | Stress-activated map kinase-interacting protein 1 | TORC2 subunit MAPKAP1 | Target of rapamycin complex 2 subunit MAPKAP1 | mSIN1 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 59129.70 |
|---|
| Organism: | Homo sapiens |
|---|
| Description: | ChEMBL_119716 |
|---|
| Residue: | 522 |
|---|
| Sequence: | MAFLDNPTIILAHIRQSHVTSDDTGMCEMVLIDHDVDLEKIHPPSMPGDSGSEIQGSNGE
TQGYVYAQSVDITSSWDFGIRRRSNTAQRLERLRKERQNQIKCKNIQWKERNSKQSAQEL
KSLFEKKSLKEKPPISGKQSILSVRLEQCPLQLNNPFNEYSKFDGKGHVGTTATKKIDVY
LPLHSSQDRLLPMTVVTMASARVQDLIGLICWQYTSEGREPKLNDNVSAYCLHIAEDDGE
VDTDFPPLDSNEPIHKFGFSTLALVEKYSSPGLTSKESLFVRINAAHGFSLIQVDNTKVT
MKEILLKAVKRRKGSQKVSGPQYRLEKQSEPNVAVDLDSTLESQSAWEFCLVRENSSRAD
GVFEEDSQIDIATVQDMLSSHHYKSFKVSMIHRLRFTTDVQLGISGDKVEIDPVTNQKAS
TKFWIKQKPISIDSDLLCACDLAEEKSPSHAIFKLTYLSNHDYKHLYFESDAATVNEIVL
KVNYILESRASTARADYFAQKQRKLNRRTSFSFQKEKKSGQQ
|
|
|
|---|
| BDBM50505422 |
|---|
| n/a |
|---|
| Name | BDBM50505422 |
|---|
| Synonyms: | CHEMBL4562100 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H26N6O2 |
|---|
| Mol. Mass. | 382.4594 |
|---|
| SMILES | [H][C@]12Cc3c(nc(nc3N3CCOCC3(C)C)-c3ccc(N)nc3)N1CCOC2 |r| |
|---|
| Structure | 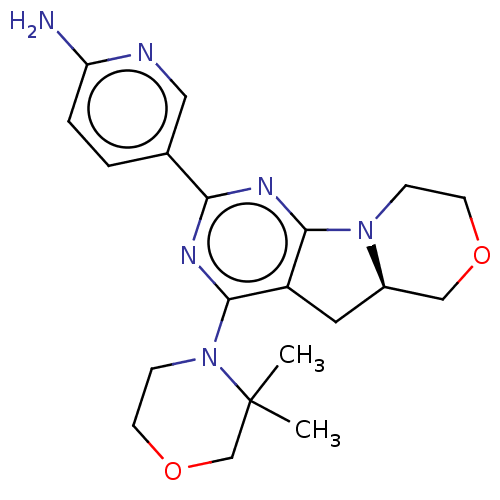
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Borsari, C; Rageot, D; Dall'Asen, A; Bohnacker, T; Melone, A; Sele, AM; Jackson, E; Langlois, JB; Beaufils, F; Hebeisen, P; Fabbro, D; Hillmann, P; Wymann, MP A Conformational Restriction Strategy for the Identification of a Highly Selective Pyrimido-pyrrolo-oxazine mTOR Inhibitor. J Med Chem62:8609-8630 (2019) [PubMed] Article
Borsari, C; Rageot, D; Dall'Asen, A; Bohnacker, T; Melone, A; Sele, AM; Jackson, E; Langlois, JB; Beaufils, F; Hebeisen, P; Fabbro, D; Hillmann, P; Wymann, MP A Conformational Restriction Strategy for the Identification of a Highly Selective Pyrimido-pyrrolo-oxazine mTOR Inhibitor. J Med Chem62:8609-8630 (2019) [PubMed] Article