| Reaction Details |
|---|
 | Report a problem with these data |
| Target | 5-hydroxytryptamine receptor 1A |
|---|
| Ligand | BDBM50069261 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_201044 |
|---|
| Ki | 125±n/a nM |
|---|
| Citation |  López-Rodríguez, ML; Morcillo, MJ; Rovat, TK; Fernández, E; Sanz, AM; Orensanz, L 1-[omega-(4-arylpiperazin-1-yl)alkyl]-3-diphenylmethylene-2,5- pyrrolidinediones as 5-HT1A receptor ligands: study of the steric requirements of the terminal amide fragment on 5-HT1A affinity/selectivity. Bioorg Med Chem Lett8:581-6 (1999) [PubMed] López-Rodríguez, ML; Morcillo, MJ; Rovat, TK; Fernández, E; Sanz, AM; Orensanz, L 1-[omega-(4-arylpiperazin-1-yl)alkyl]-3-diphenylmethylene-2,5- pyrrolidinediones as 5-HT1A receptor ligands: study of the steric requirements of the terminal amide fragment on 5-HT1A affinity/selectivity. Bioorg Med Chem Lett8:581-6 (1999) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| 5-hydroxytryptamine receptor 1A |
|---|
| Name: | 5-hydroxytryptamine receptor 1A |
|---|
| Synonyms: | 5-HT-1A | 5-HT1 | 5-HT1A | 5-Hydroxytryptamine receptor 1A (5-HT1A) | 5-hydroxytryptamine receptor 1A (5HT1A) | 5HT1A_RAT | 5ht1a | G-21 | Htr1a | Serotonin 1 (5-HT1) receptor | Serotonin 1a (5-HT1a) receptor/Adrenergic receptor alpha-1 | Serotonin receptor 1A |
|---|
| Type: | G Protein-Coupled Receptor (GPCR) |
|---|
| Mol. Mass.: | 46445.29 |
|---|
| Organism: | Rattus norvegicus (rat) |
|---|
| Description: | Binding assays were performed using rat hippocampal membranes. |
|---|
| Residue: | 422 |
|---|
| Sequence: | MDVFSFGQGNNTTASQEPFGTGGNVTSISDVTFSYQVITSLLLGTLIFCAVLGNACVVAA
IALERSLQNVANYLIGSLAVTDLMVSVLVLPMAALYQVLNKWTLGQVTCDLFIALDVLCC
TSSILHLCAIALDRYWAITDPIDYVNKRTPRRAAALISLTWLIGFLISIPPMLGWRTPED
RSDPDACTISKDHGYTIYSTFGAFYIPLLLMLVLYGRIFRAARFRIRKTVRKVEKKGAGT
SLGTSSAPPPKKSLNGQPGSGDWRRCAENRAVGTPCTNGAVRQGDDEATLEVIEVHRVGN
SKEHLPLPSESGSNSYAPACLERKNERNAEAKRKMALARERKTVKTLGIIMGTFILCWLP
FFIVALVLPFCESSCHMPALLGAIINWLGYSNSLLNPVIYAYFNKDFQNAFKKIIKCKFC
RR
|
|
|
|---|
| BDBM50069261 |
|---|
| n/a |
|---|
| Name | BDBM50069261 |
|---|
| Synonyms: | 3-Benzhydrylidene-1-[4-(4-phenyl-piperazin-1-yl)-butyl]-pyrrolidine-2,5-dione | CHEMBL342995 | cid_9511286 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C31H33N3O2 |
|---|
| Mol. Mass. | 479.6126 |
|---|
| SMILES | O=[#6]-1-[#6]\[#6](=[#6](/c2ccccc2)-c2ccccc2)-[#6](=O)-[#7]-1-[#6]-[#6]-[#6]-[#6]-[#7]-1-[#6]-[#6]-[#7](-[#6]-[#6]-1)-c1ccccc1 |
|---|
| Structure | 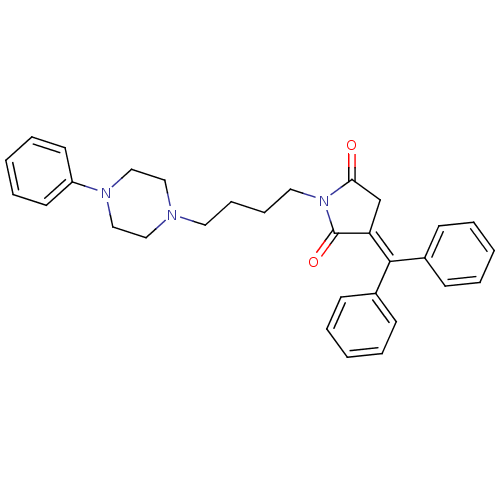
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 López-Rodríguez, ML; Morcillo, MJ; Rovat, TK; Fernández, E; Sanz, AM; Orensanz, L 1-[omega-(4-arylpiperazin-1-yl)alkyl]-3-diphenylmethylene-2,5- pyrrolidinediones as 5-HT1A receptor ligands: study of the steric requirements of the terminal amide fragment on 5-HT1A affinity/selectivity. Bioorg Med Chem Lett8:581-6 (1999) [PubMed]
López-Rodríguez, ML; Morcillo, MJ; Rovat, TK; Fernández, E; Sanz, AM; Orensanz, L 1-[omega-(4-arylpiperazin-1-yl)alkyl]-3-diphenylmethylene-2,5- pyrrolidinediones as 5-HT1A receptor ligands: study of the steric requirements of the terminal amide fragment on 5-HT1A affinity/selectivity. Bioorg Med Chem Lett8:581-6 (1999) [PubMed]