| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cytochrome P450 1A2 |
|---|
| Ligand | BDBM50529975 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1911028 (CHEMBL4413474) |
|---|
| IC50 | <700±n/a nM |
|---|
| Citation |  Graziani, D; Caligari, S; Callegari, E; De Toma, C; Longhi, M; Frigerio, F; Dilernia, R; Menegon, S; Pinzi, L; Pirona, L; Tazzari, V; Valsecchi, AE; Vistoli, G; Rastelli, G; Riva, C Evaluation of Amides, Carbamates, Sulfonamides, and Ureas of 4-Prop-2-ynylidenecycloalkylamine as Potent, Selective, and Bioavailable Negative Allosteric Modulators of Metabotropic Glutamate Receptor 5. J Med Chem62:1246-1273 (2019) [PubMed] Article Graziani, D; Caligari, S; Callegari, E; De Toma, C; Longhi, M; Frigerio, F; Dilernia, R; Menegon, S; Pinzi, L; Pirona, L; Tazzari, V; Valsecchi, AE; Vistoli, G; Rastelli, G; Riva, C Evaluation of Amides, Carbamates, Sulfonamides, and Ureas of 4-Prop-2-ynylidenecycloalkylamine as Potent, Selective, and Bioavailable Negative Allosteric Modulators of Metabotropic Glutamate Receptor 5. J Med Chem62:1246-1273 (2019) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cytochrome P450 1A2 |
|---|
| Name: | Cytochrome P450 1A2 |
|---|
| Synonyms: | CP1A2_HUMAN | CYP1A2 | CYPIA2 | Cholesterol 25-hydroxylase | Cytochrome P(3)450 | Cytochrome P450 1A | Cytochrome P450 1A2 (CYP1A2) | Cytochrome P450 4 | Cytochrome P450-P3 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 58423.38 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P05177 |
|---|
| Residue: | 516 |
|---|
| Sequence: | MALSQSVPFSATELLLASAIFCLVFWVLKGLRPRVPKGLKSPPEPWGWPLLGHVLTLGKN
PHLALSRMSQRYGDVLQIRIGSTPVLVLSRLDTIRQALVRQGDDFKGRPDLYTSTLITDG
QSLTFSTDSGPVWAARRRLAQNALNTFSIASDPASSSSCYLEEHVSKEAKALISRLQELM
AGPGHFDPYNQVVVSVANVIGAMCFGQHFPESSDEMLSLVKNTHEFVETASSGNPLDFFP
ILRYLPNPALQRFKAFNQRFLWFLQKTVQEHYQDFDKNSVRDITGALFKHSKKGPRASGN
LIPQEKIVNLVNDIFGAGFDTVTTAISWSLMYLVTKPEIQRKIQKELDTVIGRERRPRLS
DRPQLPYLEAFILETFRHSSFLPFTIPHSTTRDTTLNGFYIPKKCCVFVNQWQVNHDPEL
WEDPSEFRPERFLTADGTAINKPLSEKMMLFGMGKRRCIGEVLAKWEIFLFLAILLQQLE
FSVPPGVKVDLTPIYGLTMKHARCEHVQARLRFSIN
|
|
|
|---|
| BDBM50529975 |
|---|
| n/a |
|---|
| Name | BDBM50529975 |
|---|
| Synonyms: | CHEMBL4539998 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C16H18N2O2 |
|---|
| Mol. Mass. | 270.3263 |
|---|
| SMILES | CCOC(=O)N1CC\C(C1)=C\C#Cc1cccc(C)n1 |
|---|
| Structure | 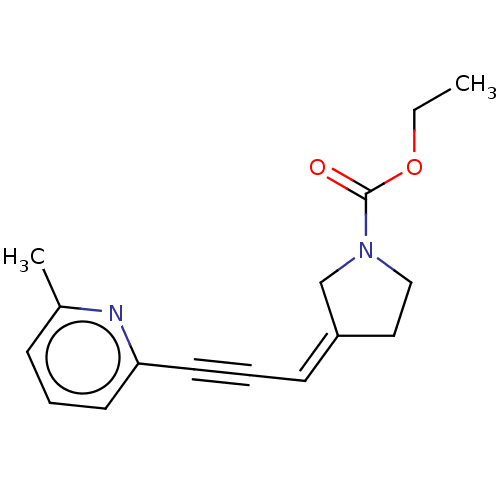
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Graziani, D; Caligari, S; Callegari, E; De Toma, C; Longhi, M; Frigerio, F; Dilernia, R; Menegon, S; Pinzi, L; Pirona, L; Tazzari, V; Valsecchi, AE; Vistoli, G; Rastelli, G; Riva, C Evaluation of Amides, Carbamates, Sulfonamides, and Ureas of 4-Prop-2-ynylidenecycloalkylamine as Potent, Selective, and Bioavailable Negative Allosteric Modulators of Metabotropic Glutamate Receptor 5. J Med Chem62:1246-1273 (2019) [PubMed] Article
Graziani, D; Caligari, S; Callegari, E; De Toma, C; Longhi, M; Frigerio, F; Dilernia, R; Menegon, S; Pinzi, L; Pirona, L; Tazzari, V; Valsecchi, AE; Vistoli, G; Rastelli, G; Riva, C Evaluation of Amides, Carbamates, Sulfonamides, and Ureas of 4-Prop-2-ynylidenecycloalkylamine as Potent, Selective, and Bioavailable Negative Allosteric Modulators of Metabotropic Glutamate Receptor 5. J Med Chem62:1246-1273 (2019) [PubMed] Article