| Reaction Details |
|---|
 | Report a problem with these data |
| Target | D(2) dopamine receptor |
|---|
| Ligand | BDBM50021365 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_61121 |
|---|
| Ki | 5.4±n/a nM |
|---|
| Citation |  Nishimura, T; Igarashi, J; Sunagawa, M Conformational analysis of tandospirone in aqueous solution: lead evolution of potent dopamine D4 receptor ligands. Bioorg Med Chem Lett11:1141-4 (2001) [PubMed] Nishimura, T; Igarashi, J; Sunagawa, M Conformational analysis of tandospirone in aqueous solution: lead evolution of potent dopamine D4 receptor ligands. Bioorg Med Chem Lett11:1141-4 (2001) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| D(2) dopamine receptor |
|---|
| Name: | D(2) dopamine receptor |
|---|
| Synonyms: | D(2) dopamine receptor | DOPAMINE D2 | DOPAMINE D2 Long | DOPAMINE D2 Short | DRD2 | DRD2_HUMAN | Dopamine D2 receptor | Dopamine D2 receptor (D2) | Dopamine D2 receptor (D2R) | Dopamine D2A | Dopamine2-like | d2 |
|---|
| Type: | Cell-surface receptors |
|---|
| Mol. Mass.: | 50647.10 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P14416 |
|---|
| Residue: | 443 |
|---|
| Sequence: | MDPLNLSWYDDDLERQNWSRPFNGSDGKADRPHYNYYATLLTLLIAVIVFGNVLVCMAVS
REKALQTTTNYLIVSLAVADLLVATLVMPWVVYLEVVGEWKFSRIHCDIFVTLDVMMCTA
SILNLCAISIDRYTAVAMPMLYNTRYSSKRRVTVMISIVWVLSFTISCPLLFGLNNADQN
ECIIANPAFVVYSSIVSFYVPFIVTLLVYIKIYIVLRRRRKRVNTKRSSRAFRAHLRAPL
KGNCTHPEDMKLCTVIMKSNGSFPVNRRRVEAARRAQELEMEMLSSTSPPERTRYSPIPP
SHHQLTLPDPSHHGLHSTPDSPAKPEKNGHAKDHPKIAKIFEIQTMPNGKTRTSLKTMSR
RKLSQQKEKKATQMLAIVLGVFIICWLPFFITHILNIHCDCNIPPVLYSAFTWLGYVNSA
VNPIIYTTFNIEFRKAFLKILHC
|
|
|
|---|
| BDBM50021365 |
|---|
| n/a |
|---|
| Name | BDBM50021365 |
|---|
| Synonyms: | CHEMBL407641 | Mezilamine | [4-Chloro-6-(4-methyl-piperazin-1-yl)-5-methylsulfanyl-pyrimidin-2-yl]-methyl-amine |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C11H18ClN5S |
|---|
| Mol. Mass. | 287.812 |
|---|
| SMILES | CNc1nc(Cl)c(SC)c(n1)N1CCN(C)CC1 |
|---|
| Structure | 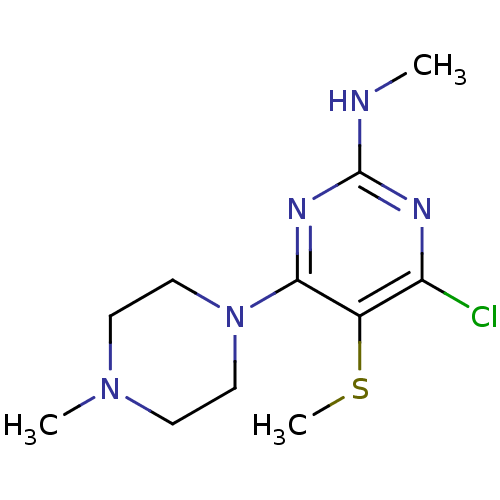
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Nishimura, T; Igarashi, J; Sunagawa, M Conformational analysis of tandospirone in aqueous solution: lead evolution of potent dopamine D4 receptor ligands. Bioorg Med Chem Lett11:1141-4 (2001) [PubMed]
Nishimura, T; Igarashi, J; Sunagawa, M Conformational analysis of tandospirone in aqueous solution: lead evolution of potent dopamine D4 receptor ligands. Bioorg Med Chem Lett11:1141-4 (2001) [PubMed]