| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Sorbitol dehydrogenase |
|---|
| Ligand | BDBM50102723 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_223912 |
|---|
| IC50 | 1000±n/a nM |
|---|
| Citation |  Darmanin, C; El-Kabbani, O Modelling studies of the active site of human sorbitol dehydrogenase: an approach to structure-based inhibitor design of the enzyme. Bioorg Med Chem Lett11:3133-6 (2001) [PubMed] Darmanin, C; El-Kabbani, O Modelling studies of the active site of human sorbitol dehydrogenase: an approach to structure-based inhibitor design of the enzyme. Bioorg Med Chem Lett11:3133-6 (2001) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Sorbitol dehydrogenase |
|---|
| Name: | Sorbitol dehydrogenase |
|---|
| Synonyms: | DHSO_HUMAN | L-iditol 2-dehydrogenase | SORD |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 38332.20 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_223910 |
|---|
| Residue: | 357 |
|---|
| Sequence: | MAAAAKPNNLSLVVHGPGDLRLENYPIPEPGPNEVLLRMHSVGICGSDVHYWEYGRIGNF
IVKKPMVLGHEASGTVEKVGSSVKHLKPGDRVAIEPGAPRENDEFCKMGRYNLSPSIFFC
ATPPDDGNLCRFYKHNAAFCYKLPDNVTFEEGALIEPLSVGIHACRRGGVTLGHKVLVCG
AGPIGMVTLLVAKAMGAAQVVVTDLSATRLSKAKEIGADLVLQISKESPQEIARKVEGQL
GCKPEVTIECTGAEASIQAGIYATRSGGNLVLVGLGSEMTTVPLLHAAIREVDIKGVFRY
CNTWPVAISMLASKSVNVKPLVTHRFPLEKALEAFETFKKGLGLKIMLKCDPSDQNP
|
|
|
|---|
| BDBM50102723 |
|---|
| n/a |
|---|
| Name | BDBM50102723 |
|---|
| Synonyms: | 4-(2-Hydroxymethyl-pyrimidin-4-yl)-piperazine-1-sulfonic acid dimethylamide | 4-[2-(HYDROXYMETHYL)PYRIMIDIN-4-YL]-N,N-DIMETHYLPIPERAZINE-1-SULFONAMIDE | CHEMBL90488 | CP-166572 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C11H19N5O3S |
|---|
| Mol. Mass. | 301.365 |
|---|
| SMILES | CN(C)S(=O)(=O)N1CCN(CC1)c1ccnc(CO)n1 |
|---|
| Structure | 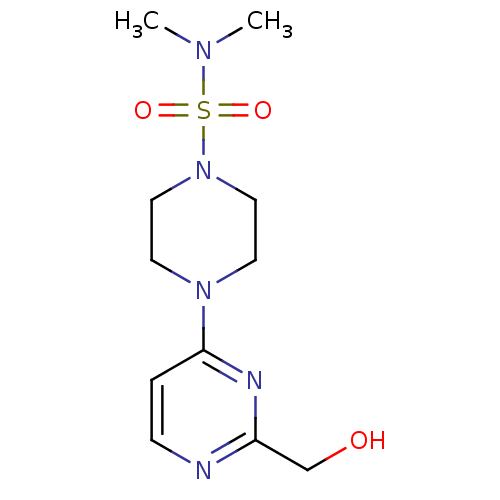
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Darmanin, C; El-Kabbani, O Modelling studies of the active site of human sorbitol dehydrogenase: an approach to structure-based inhibitor design of the enzyme. Bioorg Med Chem Lett11:3133-6 (2001) [PubMed]
Darmanin, C; El-Kabbani, O Modelling studies of the active site of human sorbitol dehydrogenase: an approach to structure-based inhibitor design of the enzyme. Bioorg Med Chem Lett11:3133-6 (2001) [PubMed]