| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Mucosa-associated lymphoid tissue lymphoma translocation protein 1 |
|---|
| Ligand | BDBM50569466 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2109369 (CHEMBL4818044) |
|---|
| IC50 | 21180±n/a nM |
|---|
| Citation |  Asaba, KN; Adachi, Y; Tokumaru, K; Watanabe, A; Goto, Y; Aoki, T Structure-activity relationship studies of 3-substituted pyrazoles as novel allosteric inhibitors of MALT1 protease. Bioorg Med Chem Lett41:0 (2021) [PubMed] Article Asaba, KN; Adachi, Y; Tokumaru, K; Watanabe, A; Goto, Y; Aoki, T Structure-activity relationship studies of 3-substituted pyrazoles as novel allosteric inhibitors of MALT1 protease. Bioorg Med Chem Lett41:0 (2021) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Mucosa-associated lymphoid tissue lymphoma translocation protein 1 |
|---|
| Name: | Mucosa-associated lymphoid tissue lymphoma translocation protein 1 |
|---|
| Synonyms: | MALT lymphoma-associated translocation | MALT lymphoma-associated translocation (MALT1) | MALT1 | MALT1_HUMAN | MLT | Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | Paracaspase |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 92257.81 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9UDY8 |
|---|
| Residue: | 824 |
|---|
| Sequence: | MSLLGDPLQALPPSAAPTGPLLAPPAGATLNRLREPLLRRLSELLDQAPEGRGWRRLAEL
AGSRGRLRLSCLDLEQCSLKVLEPEGSPSLCLLKLMGEKGCTVTELSDFLQAMEHTEVLQ
LLSPPGIKITVNPESKAVLAGQFVKLCCRATGHPFVQYQWFKMNKEIPNGNTSELIFNAV
HVKDAGFYVCRVNNNFTFEFSQWSQLDVCDIPESFQRSVDGVSESKLQICVEPTSQKLMP
GSTLVLQCVAVGSPIPHYQWFKNELPLTHETKKLYMVPYVDLEHQGTYWCHVYNDRDSQD
SKKVEIIIGRTDEAVECTEDELNNLGHPDNKEQTTDQPLAKDKVALLIGNMNYREHPKLK
APLVDVYELTNLLRQLDFKVVSLLDLTEYEMRNAVDEFLLLLDKGVYGLLYYAGHGYENF
GNSFMVPVDAPNPYRSENCLCVQNILKLMQEKETGLNVFLLDMCRKRNDYDDTIPILDAL
KVTANIVFGYATCQGAEAFEIQHSGLANGIFMKFLKDRLLEDKKITVLLDEVAEDMGKCH
LTKGKQALEIRSSLSEKRALTDPIQGTEYSAESLVRNLQWAKAHELPESMCLKFDCGVQI
QLGFAAEFSNVMIIYTSIVYKPPEIIMCDAYVTDFPLDLDIDPKDANKGTPEETGSYLVS
KDLPKHCLYTRLSSLQKLKEHLVFTVCLSYQYSGLEDTVEDKQEVNVGKPLIAKLDMHRG
LGRKTCFQTCLMSNGPYQSSAATSGGAGHYHSLQDPFHGVYHSHPGNPSNVTPADSCHCS
RTPDAFISSFAHHASCHFSRSNVPVETTDEIPFSFSDRLRISEK
|
|
|
|---|
| BDBM50569466 |
|---|
| n/a |
|---|
| Name | BDBM50569466 |
|---|
| Synonyms: | CHEMBL3972284 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H15Cl2N3O |
|---|
| Mol. Mass. | 360.237 |
|---|
| SMILES | CN(C)C(=O)c1cc(-c2ccc(Cl)cc2)n(n1)-c1ccc(Cl)cc1 |
|---|
| Structure | 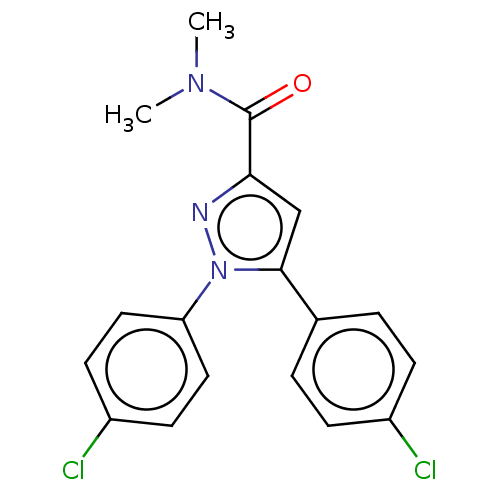
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Asaba, KN; Adachi, Y; Tokumaru, K; Watanabe, A; Goto, Y; Aoki, T Structure-activity relationship studies of 3-substituted pyrazoles as novel allosteric inhibitors of MALT1 protease. Bioorg Med Chem Lett41:0 (2021) [PubMed] Article
Asaba, KN; Adachi, Y; Tokumaru, K; Watanabe, A; Goto, Y; Aoki, T Structure-activity relationship studies of 3-substituted pyrazoles as novel allosteric inhibitors of MALT1 protease. Bioorg Med Chem Lett41:0 (2021) [PubMed] Article