| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Mitogen-activated protein kinase 3 |
|---|
| Ligand | BDBM50315615 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_623754 (CHEMBL1114419) |
|---|
| IC50 | 980±n/a nM |
|---|
| Citation |  Blum, CA; Caldwell, T; Zheng, X; Bakthavatchalam, R; Capitosti, S; Brielmann, H; De Lombaert, S; Kershaw, MT; Matson, D; Krause, JE; Cortright, D; Crandall, M; Martin, WJ; Murphy, BA; Boyce, S; Jones, AB; Mason, G; Rycroft, W; Perrett, H; Conley, R; Burnaby-Davies, N; Chenard, BL; Hodgetts, KJ Discovery of novel 6,6-heterocycles as transient receptor potential vanilloid (TRPV1) antagonists. J Med Chem53:3330-48 (2010) [PubMed] Article Blum, CA; Caldwell, T; Zheng, X; Bakthavatchalam, R; Capitosti, S; Brielmann, H; De Lombaert, S; Kershaw, MT; Matson, D; Krause, JE; Cortright, D; Crandall, M; Martin, WJ; Murphy, BA; Boyce, S; Jones, AB; Mason, G; Rycroft, W; Perrett, H; Conley, R; Burnaby-Davies, N; Chenard, BL; Hodgetts, KJ Discovery of novel 6,6-heterocycles as transient receptor potential vanilloid (TRPV1) antagonists. J Med Chem53:3330-48 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Mitogen-activated protein kinase 3 |
|---|
| Name: | Mitogen-activated protein kinase 3 |
|---|
| Synonyms: | ERK1 | ERK1/ERK2 | Extracellular signal-regulated kinase 1 | Extracellular signal-regulated kinase 1 (ERK1) | MAP kinase ERK1 | MAPK3 | MAPK3/MAPK7 | MK03_HUMAN | Mitogen-activated protein kinase | Mitogen-activated protein kinase 3 | PRKM3 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 43136.58 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | curated by PDB 4QTB |
|---|
| Residue: | 379 |
|---|
| Sequence: | MAAAAAQGGGGGEPRRTEGVGPGVPGEVEMVKGQPFDVGPRYTQLQYIGEGAYGMVSSAY
DHVRKTRVAIKKISPFEHQTYCQRTLREIQILLRFRHENVIGIRDILRASTLEAMRDVYI
VQDLMETDLYKLLKSQQLSNDHICYFLYQILRGLKYIHSANVLHRDLKPSNLLINTTCDL
KICDFGLARIADPEHDHTGFLTEYVATRWYRAPEIMLNSKGYTKSIDIWSVGCILAEMLS
NRPIFPGKHYLDQLNHILGILGSPSQEDLNCIINMKARNYLQSLPSKTKVAWAKLFPKSD
SKALDLLDRMLTFNPNKRITVEEALAHPYLEQYYDPTDEPVAEEPFTFAMELDDLPKERL
KELIFQETARFQPGVLEAP
|
|
|
|---|
| BDBM50315615 |
|---|
| n/a |
|---|
| Name | BDBM50315615 |
|---|
| Synonyms: | 7-(3-(trifluoromethyl)pyridin-2-yl)-N-(5-(trifluoromethyl)pyridin-2-yl)-1,8-naphthyridin-4-amine | 7-[3-(Trifluoromethyl)pyridin-2-yl]-N-[5-(trifluoromethyl)-pyridin-2-yl]-1,8-naphthyridin-4-amine | CHEMBL1089119 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H11F6N5 |
|---|
| Mol. Mass. | 435.3253 |
|---|
| SMILES | FC(F)(F)c1ccc(Nc2ccnc3nc(ccc23)-c2ncccc2C(F)(F)F)nc1 |
|---|
| Structure | 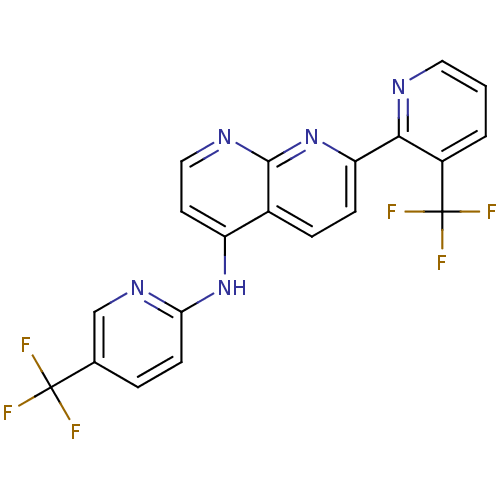
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Blum, CA; Caldwell, T; Zheng, X; Bakthavatchalam, R; Capitosti, S; Brielmann, H; De Lombaert, S; Kershaw, MT; Matson, D; Krause, JE; Cortright, D; Crandall, M; Martin, WJ; Murphy, BA; Boyce, S; Jones, AB; Mason, G; Rycroft, W; Perrett, H; Conley, R; Burnaby-Davies, N; Chenard, BL; Hodgetts, KJ Discovery of novel 6,6-heterocycles as transient receptor potential vanilloid (TRPV1) antagonists. J Med Chem53:3330-48 (2010) [PubMed] Article
Blum, CA; Caldwell, T; Zheng, X; Bakthavatchalam, R; Capitosti, S; Brielmann, H; De Lombaert, S; Kershaw, MT; Matson, D; Krause, JE; Cortright, D; Crandall, M; Martin, WJ; Murphy, BA; Boyce, S; Jones, AB; Mason, G; Rycroft, W; Perrett, H; Conley, R; Burnaby-Davies, N; Chenard, BL; Hodgetts, KJ Discovery of novel 6,6-heterocycles as transient receptor potential vanilloid (TRPV1) antagonists. J Med Chem53:3330-48 (2010) [PubMed] Article