| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Poly [ADP-ribose] polymerase tankyrase-2 |
|---|
| Ligand | BDBM50294816 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_818396 (CHEMBL2034623) |
|---|
| EC50 | 10000±n/a nM |
|---|
| Citation |  Narwal, M; Venkannagari, H; Lehtiö, L Structural basis of selective inhibition of human tankyrases. J Med Chem55:1360-7 (2012) [PubMed] Article Narwal, M; Venkannagari, H; Lehtiö, L Structural basis of selective inhibition of human tankyrases. J Med Chem55:1360-7 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Poly [ADP-ribose] polymerase tankyrase-2 |
|---|
| Name: | Poly [ADP-ribose] polymerase tankyrase-2 |
|---|
| Synonyms: | (ARTD6 or PARP5b) | PARP5B | Poly [ADP-ribose] polymerase tankyrase-2 | TANK2 | TNKL | TNKS2 | TNKS2_HUMAN | TPoly [ADP-ribose] polymerase tankyrase-2 | Tankyrase 2 | Tankyrase II | Tankyrase-2 | Tankyrase-2 (TNKS-2) | Tankyrase-2 (TNKS2) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 126937.16 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9H2K2 |
|---|
| Residue: | 1166 |
|---|
| Sequence: | MSGRRCAGGGAACASAAAEAVEPAARELFEACRNGDVERVKRLVTPEKVNSRDTAGRKST
PLHFAAGFGRKDVVEYLLQNGANVQARDDGGLIPLHNACSFGHAEVVNLLLRHGADPNAR
DNWNYTPLHEAAIKGKIDVCIVLLQHGAEPTIRNTDGRTALDLADPSAKAVLTGEYKKDE
LLESARSGNEEKMMALLTPLNVNCHASDGRKSTPLHLAAGYNRVKIVQLLLQHGADVHAK
DKGDLVPLHNACSYGHYEVTELLVKHGACVNAMDLWQFTPLHEAASKNRVEVCSLLLSYG
ADPTLLNCHNKSAIDLAPTPQLKERLAYEFKGHSLLQAAREADVTRIKKHLSLEMVNFKH
PQTHETALHCAAASPYPKRKQICELLLRKGANINEKTKEFLTPLHVASEKAHNDVVEVVV
KHEAKVNALDNLGQTSLHRAAYCGHLQTCRLLLSYGCDPNIISLQGFTALQMGNENVQQL
LQEGISLGNSEADRQLLEAAKAGDVETVKKLCTVQSVNCRDIEGRQSTPLHFAAGYNRVS
VVEYLLQHGADVHAKDKGGLVPLHNACSYGHYEVAELLVKHGAVVNVADLWKFTPLHEAA
AKGKYEICKLLLQHGADPTKKNRDGNTPLDLVKDGDTDIQDLLRGDAALLDAAKKGCLAR
VKKLSSPDNVNCRDTQGRHSTPLHLAAGYNNLEVAEYLLQHGADVNAQDKGGLIPLHNAA
SYGHVDVAALLIKYNACVNATDKWAFTPLHEAAQKGRTQLCALLLAHGADPTLKNQEGQT
PLDLVSADDVSALLTAAMPPSALPSCYKPQVLNGVRSPGATADALSSGPSSPSSLSAASS
LDNLSGSFSELSSVVSSSGTEGASSLEKKEVPGVDFSITQFVRNLGLEHLMDIFEREQIT
LDVLVEMGHKELKEIGINAYGHRHKLIKGVERLISGQQGLNPYLTLNTSGSGTILIDLSP
DDKEFQSVEEEMQSTVREHRDGGHAGGIFNRYNILKIQKVCNKKLWERYTHRRKEVSEEN
HNHANERMLFHGSPFVNAIIHKGFDERHAYIGGMFGAGIYFAENSSKSNQYVYGIGGGTG
CPVHKDRSCYICHRQLLFCRVTLGKSFLQFSAMKMAHSPPGHHSVTGRPSVNGLALAEYV
IYRGEQAYPEYLITYQIMRPEGMVDG
|
|
|
|---|
| BDBM50294816 |
|---|
| n/a |
|---|
| Name | BDBM50294816 |
|---|
| Synonyms: | 4-((1R,2S,6R,7S)-3,5-Dioxo-4-aza-tricyclo[5.2.1.0*2,6*]dec-8-en-4-yl)-N-pyridin-4-ylmethyl-benzamide | CHEMBL551775 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H19N3O3 |
|---|
| Mol. Mass. | 373.4046 |
|---|
| SMILES | Oc1c2[C@H]3C[C@H](C=C3)c2c(O)n1-c1ccc(cc1)C(=O)NCc1ccncc1 |r,c:6| |
|---|
| Structure | 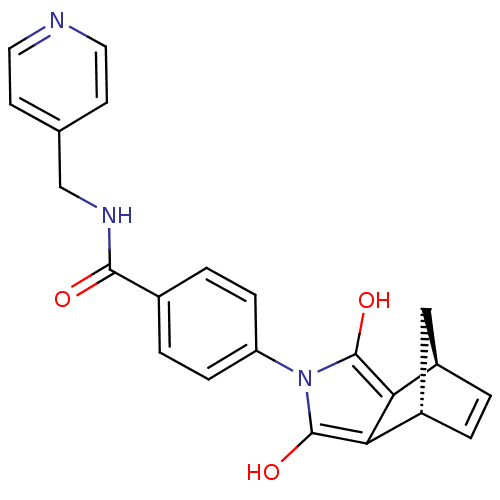
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Narwal, M; Venkannagari, H; Lehtiö, L Structural basis of selective inhibition of human tankyrases. J Med Chem55:1360-7 (2012) [PubMed] Article
Narwal, M; Venkannagari, H; Lehtiö, L Structural basis of selective inhibition of human tankyrases. J Med Chem55:1360-7 (2012) [PubMed] Article