| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Poly [ADP-ribose] polymerase 2 |
|---|
| Ligand | BDBM50444543 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1590057 (CHEMBL3829027) |
|---|
| IC50 | 10529±n/a nM |
|---|
| Citation |  Nathubhai, A; Haikarainen, T; Hayward, PC; Mu˝oz-Descalzo, S; Thompson, AS; Lloyd, MD; Lehti÷, L; Threadgill, MD Structure-activity relationships of 2-arylquinazolin-4-ones as highly selective and potent inhibitors of the tankyrases. Eur J Med Chem118:316-27 (2016) [PubMed] Article Nathubhai, A; Haikarainen, T; Hayward, PC; Mu˝oz-Descalzo, S; Thompson, AS; Lloyd, MD; Lehti÷, L; Threadgill, MD Structure-activity relationships of 2-arylquinazolin-4-ones as highly selective and potent inhibitors of the tankyrases. Eur J Med Chem118:316-27 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Poly [ADP-ribose] polymerase 2 |
|---|
| Name: | Poly [ADP-ribose] polymerase 2 |
|---|
| Synonyms: | (ARTD2 or PARP2) | ADPRT2 | ADPRTL2 | PARP2 | PARP2_HUMAN | Poly [ADP-ribose] polymerase 2 (PARP-2) | Poly [ADP-ribose] polymerase 2 (PARP2) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 66225.70 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9UGN5 |
|---|
| Residue: | 583 |
|---|
| Sequence: | MAARRRRSTGGGRARALNESKRVNNGNTAPEDSSPAKKTRRCQRQESKKMPVAGGKANKD
RTEDKQDGMPGRSWASKRVSESVKALLLKGKAPVDPECTAKVGKAHVYCEGNDVYDVMLN
QTNLQFNNNKYYLIQLLEDDAQRNFSVWMRWGRVGKMGQHSLVACSGNLNKAKEIFQKKF
LDKTKNNWEDREKFEKVPGKYDMLQMDYATNTQDEEETKKEESLKSPLKPESQLDLRVQE
LIKLICNVQAMEEMMMEMKYNTKKAPLGKLTVAQIKAGYQSLKKIEDCIRAGQHGRALME
ACNEFYTRIPHDFGLRTPPLIRTQKELSEKIQLLEALGDIEIAIKLVKTELQSPEHPLDQ
HYRNLHCALRPLDHESYEFKVISQYLQSTHAPTHSDYTMTLLDLFEVEKDGEKEAFREDL
HNRMLLWHGSRMSNWVGILSHGLRIAPPEAPITGYMFGKGIYFADMSSKSANYCFASRLK
NTGLLLLSEVALGQCNELLEANPKAEGLLQGKHSTKGLGKMAPSSAHFVTLNGSTVPLGP
ASDTGILNPDGYTLNYNEYIVYNPNQVRMRYLLKVQFNFLQLW
|
|
|
|---|
| BDBM50444543 |
|---|
| n/a |
|---|
| Name | BDBM50444543 |
|---|
| Synonyms: | CHEMBL3098937 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C15H11BrN2O |
|---|
| Mol. Mass. | 315.165 |
|---|
| SMILES | Cc1cccc2c1nc([nH]c2=O)-c1ccc(Br)cc1 |
|---|
| Structure | 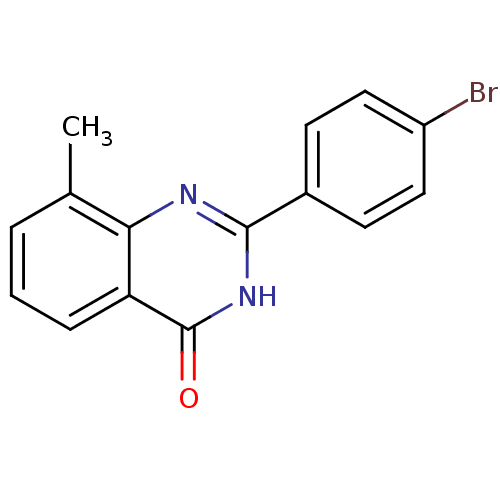
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Nathubhai, A; Haikarainen, T; Hayward, PC; Mu˝oz-Descalzo, S; Thompson, AS; Lloyd, MD; Lehti÷, L; Threadgill, MD Structure-activity relationships of 2-arylquinazolin-4-ones as highly selective and potent inhibitors of the tankyrases. Eur J Med Chem118:316-27 (2016) [PubMed] Article
Nathubhai, A; Haikarainen, T; Hayward, PC; Mu˝oz-Descalzo, S; Thompson, AS; Lloyd, MD; Lehti÷, L; Threadgill, MD Structure-activity relationships of 2-arylquinazolin-4-ones as highly selective and potent inhibitors of the tankyrases. Eur J Med Chem118:316-27 (2016) [PubMed] Article