| Reaction Details |
|---|
 | Report a problem with these data |
| Target | cAMP-specific 3',5'-cyclic phosphodiesterase 4A |
|---|
| Ligand | BDBM285602 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Scintillation Proximity Assay (SPA) |
|---|
| IC50 | 10.5±n/a nM |
|---|
| Citation |  Chappie, TA; Verhoest, PR; Patel, NC; Hayward, MM; Helal, CJ; Sciabola, S; LaChapelle, EA; Young, JM Imidazopyridazine compounds US Patent US9598421 Publication Date 3/21/2017 Chappie, TA; Verhoest, PR; Patel, NC; Hayward, MM; Helal, CJ; Sciabola, S; LaChapelle, EA; Young, JM Imidazopyridazine compounds US Patent US9598421 Publication Date 3/21/2017 |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| cAMP-specific 3',5'-cyclic phosphodiesterase 4A |
|---|
| Name: | cAMP-specific 3',5'-cyclic phosphodiesterase 4A |
|---|
| Synonyms: | 3',5'-cyclic phosphodiesterase | DPDE2 | PDE46 | PDE4A | PDE4A_HUMAN | Phosphodiesterase 4 (PDE4) | Phosphodiesterase 4A | Phosphodiesterase 4A (PDE4) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 98113.27 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P27815 |
|---|
| Residue: | 886 |
|---|
| Sequence: | MEPPTVPSERSLSLSLPGPREGQATLKPPPQHLWRQPRTPIRIQQRGYSDSAERAERERQ
PHRPIERADAMDTSDRPGLRTTRMSWPSSFHGTGTGSGGAGGGSSRRFEAENGPTPSPGR
SPLDSQASPGLVLHAGAATSQRRESFLYRSDSDYDMSPKTMSRNSSVTSEAHAEDLIVTP
FAQVLASLRSVRSNFSLLTNVPVPSNKRSPLGGPTPVCKATLSEETCQQLARETLEELDW
CLEQLETMQTYRSVSEMASHKFKRMLNRELTHLSEMSRSGNQVSEYISTTFLDKQNEVEI
PSPTMKEREKQQAPRPRPSQPPPPPVPHLQPMSQITGLKKLMHSNSLNNSNIPRFGVKTD
QEELLAQELENLNKWGLNIFCVSDYAGGRSLTCIMYMIFQERDLLKKFRIPVDTMVTYML
TLEDHYHADVAYHNSLHAADVLQSTHVLLATPALDAVFTDLEILAALFAAAIHDVDHPGV
SNQFLINTNSELALMYNDESVLENHHLAVGFKLLQEDNCDIFQNLSKRQRQSLRKMVIDM
VLATDMSKHMTLLADLKTMVETKKVTSSGVLLLDNYSDRIQVLRNMVHCADLSNPTKPLE
LYRQWTDRIMAEFFQQGDRERERGMEISPMCDKHTASVEKSQVGFIDYIVHPLWETWADL
VHPDAQEILDTLEDNRDWYYSAIRQSPSPPPEEESRGPGHPPLPDKFQFELTLEEEEEEE
ISMAQIPCTAQEALTAQGLSGVEEALDATIAWEASPAQESLEVMAQEASLEAELEAVYLT
QQAQSTGSAPVAPDEFSSREEFVVAVSHSSPSALALQSPLLPAWRTLSVSEHAPGLPGLP
STAAEVEAQREHQAAKRACSACAGTFGEDTSALPAPGGGGSGGDPT
|
|
|
|---|
| BDBM285602 |
|---|
| n/a |
|---|
| Name | BDBM285602 |
|---|
| Synonyms: | N-cyclopropyl-3-[2-(difluoro-methoxy)pyridin-4-yl]imidazo[1,2-b]pyridazine-2-carboxamide | US10077269, Example 75 | US10669279, Example 75 | US9598421, Example 75 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C16H13F2N5O2 |
|---|
| Mol. Mass. | 345.3035 |
|---|
| SMILES | FC(F)Oc1cc(ccn1)-c1c(nc2cccnn12)C(=O)NC1CC1 |
|---|
| Structure | 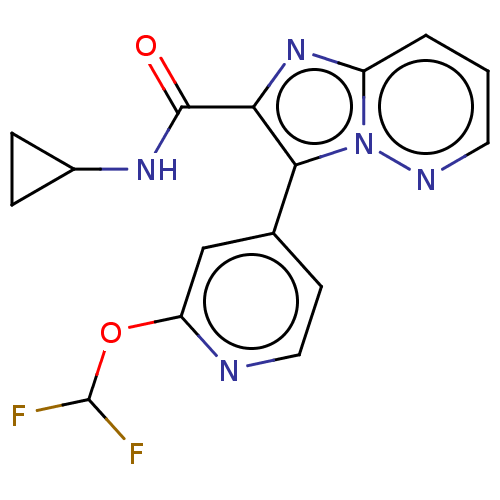
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Chappie, TA; Verhoest, PR; Patel, NC; Hayward, MM; Helal, CJ; Sciabola, S; LaChapelle, EA; Young, JM Imidazopyridazine compounds US Patent US9598421 Publication Date 3/21/2017
Chappie, TA; Verhoest, PR; Patel, NC; Hayward, MM; Helal, CJ; Sciabola, S; LaChapelle, EA; Young, JM Imidazopyridazine compounds US Patent US9598421 Publication Date 3/21/2017