| Synonyms: | PI3-kinase p110 subunit gamma | PI3-kinase subunit p120-gamma | PI3Kgamma | PIK3CG | PK3CG_HUMAN | Phosphatidylinositol 4,5-biphosphate 3-kinase catalytic subunit gamma (PIK3CG) | Phosphatidylinositol 4,5-bisphosphate 3-kinase (PI3K) | Phosphatidylinositol 4,5-bisphosphate 3-kinase 110 kDa catalytic subunit gamma (PI3K gamma) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma (PI3Kgamma) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (PI3K gamma) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (PI3K) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (PI3Kgamma) | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | Phosphoinositide 3-Kinase (PI3K), gamma Chain A | Phosphoinositide 3-kinases gamma (PI3K gamma) | Phosphoinositide-3-kinase (PI3K gamma) | p120-PI3K |
|---|
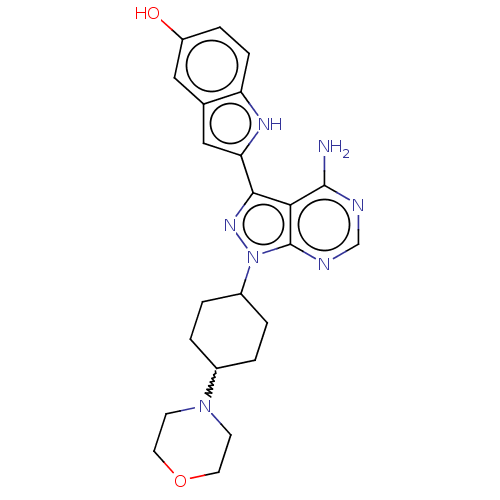
| SMILES | Nc1ncnc2n(nc(-c3cc4cc(O)ccc4[nH]3)c12)C1CCC(CC1)N1CCOCC1 |w:23.30,(-2.24,3.7,;-2.24,2.16,;-3.57,1.39,;-3.57,-.15,;-2.24,-.92,;-.9,-.15,;.56,-.63,;1.47,.62,;.56,1.86,;.96,3.35,;.06,4.6,;.96,5.84,;.64,7.35,;1.79,8.38,;1.39,9.87,;3.25,7.9,;3.57,6.4,;2.43,5.37,;2.43,3.83,;-.9,1.39,;.56,-2.17,;-.77,-2.94,;-.77,-4.48,;.56,-5.25,;1.9,-4.48,;1.9,-2.94,;.56,-6.79,;-.77,-7.56,;-.77,-9.1,;.56,-9.87,;1.9,-9.1,;1.9,-7.56,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Liu, Y; Ren, P; Jessen, K; Guo, X; Rommel, C Combination pharmaceutical compositions and uses thereof US Patent US10172858 Publication Date 1/8/2019
Liu, Y; Ren, P; Jessen, K; Guo, X; Rommel, C Combination pharmaceutical compositions and uses thereof US Patent US10172858 Publication Date 1/8/2019