| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Mitogen-activated protein kinase 14 |
|---|
| Ligand | BDBM14834 |
|---|
| Substrate/Competitor | BDBM14832 |
|---|
| Meas. Tech. | Fluorescence Exchange Curve Assay |
|---|
| pH | 7±n/a |
|---|
| Temperature | 296.15±n/a K |
|---|
| Kd | 36±n/a nM |
|---|
| Citation |  Kroe, RR; Regan, J; Proto, A; Peet, GW; Roy, T; Landro, LD; Fuschetto, NG; Pargellis, CA; Ingraham, RH Thermal denaturation: a method to rank slow binding, high-affinity P38alpha MAP kinase inhibitors. J Med Chem46:4669-75 (2003) [PubMed] Article Kroe, RR; Regan, J; Proto, A; Peet, GW; Roy, T; Landro, LD; Fuschetto, NG; Pargellis, CA; Ingraham, RH Thermal denaturation: a method to rank slow binding, high-affinity P38alpha MAP kinase inhibitors. J Med Chem46:4669-75 (2003) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Mitogen-activated protein kinase 14 |
|---|
| Name: | Mitogen-activated protein kinase 14 |
|---|
| Synonyms: | CSAID-binding protein | CSBP | CSBP1 | CSBP2 | CSPB1 | Cytokine suppressive anti-inflammatory drug-binding protein | MAP kinase 14 | MAP kinase MXI2 | MAP kinase p38 alpha | MAPK 14 | MAPK14 | MAX-interacting protein 2 | MK14_HUMAN | MXI2 | Mitogen-activated protein kinase p38 alpha | SAPK2A | Stress-activated protein kinase 2a | p38 MAP kinase alpha/beta |
|---|
| Type: | Serine/threonine-protein kinase |
|---|
| Mol. Mass.: | 41286.76 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q16539 |
|---|
| Residue: | 360 |
|---|
| Sequence: | MSQERPTFYRQELNKTIWEVPERYQNLSPVGSGAYGSVCAAFDTKTGLRVAVKKLSRPFQ
SIIHAKRTYRELRLLKHMKHENVIGLLDVFTPARSLEEFNDVYLVTHLMGADLNNIVKCQ
KLTDDHVQFLIYQILRGLKYIHSADIIHRDLKPSNLAVNEDCELKILDFGLARHTDDEMT
GYVATRWYRAPEIMLNWMHYNQTVDIWSVGCIMAELLTGRTLFPGTDHIDQLKLILRLVG
TPGAELLKKISSESARNYIQSLTQMPKMNFANVFIGANPLAVDLLEKMLVLDSDKRITAA
QALAHAYFAQYHDPDDEPVADPYDQSFESRDLLIDEWKSLTYDEVISFVPPPLDQEEMES
|
|
|
|---|
| BDBM14834 |
|---|
| BDBM14832 |
|---|
| Name | BDBM14834 |
|---|
| Synonyms: | 1-[5-(2-Hydroxy-1,1-dimethylethyl)-2-p-tolyl-2H-pyrazol-3-yl]-3-[4-(2-morpholin-4-yl-ethoxy)-naphthalen-1-yl]-urea | 3-[3-(1-hydroxy-2-methylpropan-2-yl)-1-(4-methylphenyl)-1H-pyrazol-5-yl]-1-{4-[2-(morpholin-4-yl)ethoxy]naphthalen-1-yl}urea | BIRB-796 Analog 11 | CHEMBL107850 | Dopamine D2 receptor and serotonin 2a receptor | diaryl urea compound 3 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C31H37N5O4 |
|---|
| Mol. Mass. | 543.6566 |
|---|
| SMILES | Cc1ccc(cc1)-n1nc(cc1NC(=O)Nc1ccc(OCCN2CCOCC2)c2ccccc12)C(C)(C)CO |
|---|
| Structure | 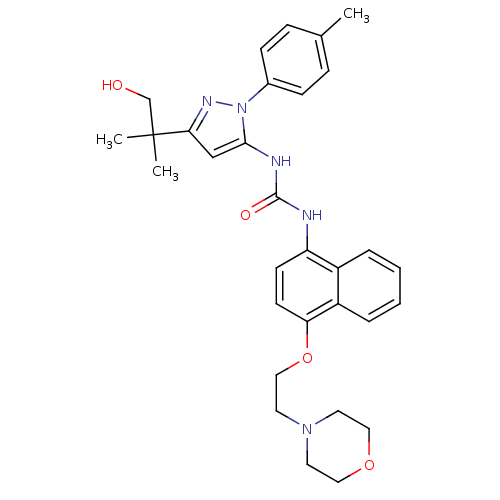
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Kroe, RR; Regan, J; Proto, A; Peet, GW; Roy, T; Landro, LD; Fuschetto, NG; Pargellis, CA; Ingraham, RH Thermal denaturation: a method to rank slow binding, high-affinity P38alpha MAP kinase inhibitors. J Med Chem46:4669-75 (2003) [PubMed] Article
Kroe, RR; Regan, J; Proto, A; Peet, GW; Roy, T; Landro, LD; Fuschetto, NG; Pargellis, CA; Ingraham, RH Thermal denaturation: a method to rank slow binding, high-affinity P38alpha MAP kinase inhibitors. J Med Chem46:4669-75 (2003) [PubMed] Article