| Reaction Details |
|---|
 | Report a problem with these data |
| Target | cGMP-specific 3',5'-cyclic phosphodiesterase [535-860] |
|---|
| Ligand | BDBM15336 |
|---|
| Substrate/Competitor | BDBM14391 |
|---|
| Meas. Tech. | Enzymatic Assay |
|---|
| pH | 7.8±n/a |
|---|
| Temperature | 297.15±n/a K |
|---|
| IC50 | 2100±500 nM |
|---|
| Km | 5100±1300 nM |
|---|
| kcat | 1.3±0.3 1/sec |
|---|
| Citation |  Wang, H; Liu, Y; Huai, Q; Cai, J; Zoraghi, R; Francis, SH; Corbin, JD; Robinson, H; Xin, Z; Lin, G; Ke, H Multiple conformations of phosphodiesterase-5: implications for enzyme function and drug development. J Biol Chem281:21469-79 (2006) [PubMed] Article Wang, H; Liu, Y; Huai, Q; Cai, J; Zoraghi, R; Francis, SH; Corbin, JD; Robinson, H; Xin, Z; Lin, G; Ke, H Multiple conformations of phosphodiesterase-5: implications for enzyme function and drug development. J Biol Chem281:21469-79 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| cGMP-specific 3',5'-cyclic phosphodiesterase [535-860] |
|---|
| Name: | cGMP-specific 3',5'-cyclic phosphodiesterase [535-860] |
|---|
| Synonyms: | CGB-PDE | PDE5 | PDE5A | PDE5A_HUMAN | Phosphodiesterase Type 5 (PDE5A) | cGMP-binding cGMP-specific phosphodiesterase | cGMP-specific 3,5-cyclic phosphodiesterase |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 37741.15 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | The coding regions for amino acids 535-860 of PDE5A1 were cloned and expressed in E. coli. |
|---|
| Residue: | 326 |
|---|
| Sequence: | EETRELQSLAAAVVPSAQTLKITDFSFSDFELSDLETALCTIRMFTDLNLVQNFQMKHEV
LCRWILSVKKNYRKNVAYHNWRHAFNTAQCMFAALKAGKIQNKLTDLEILALLIAALSHD
LDHRGVNNSYIQRSEHPLAQLYCHSIMEHHHFDQCLMILNSPGNQILSGLSIEEYKTTLK
IIKQAILATDLALYIKRRGEFFELIRKNQFNLEDPHQKELFLAMLMTACDLSAITKPWPI
QQRIAELVATEFFDQGDRERKELNIEPTDLMNREKKNKIPSMQVGFIDAICLQLYEALTH
VSEDCFPLLDGCRKNRQKWQALAEQQ
|
|
|
|---|
| BDBM15336 |
|---|
| BDBM14391 |
|---|
| Name | BDBM15336 |
|---|
| Synonyms: | 1-methyl-3-(2-methylpropyl)-2,3,6,7-tetrahydro-1H-purine-2,6-dione | 1-methyl-3-(2-methylpropyl)-3,7-dihydro-1H-purine-2,6-dione | 3-Isobutyl-1-methylxanthine | CHEMBL275084 | IBMX | isobutylmethylxanthine |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C10H14N4O2 |
|---|
| Mol. Mass. | 222.2438 |
|---|
| SMILES | CC(C)Cn1c2nc[nH]c2c(=O)n(C)c1=O |
|---|
| Structure | 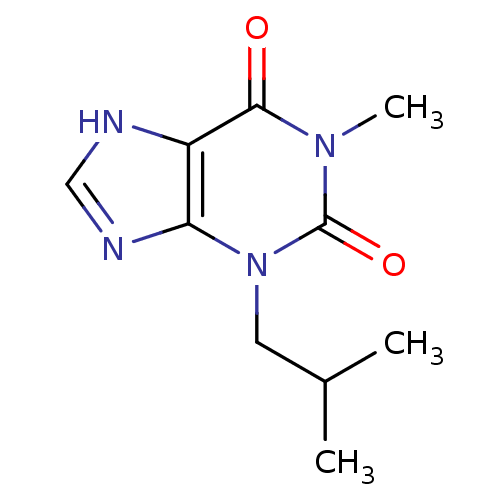
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wang, H; Liu, Y; Huai, Q; Cai, J; Zoraghi, R; Francis, SH; Corbin, JD; Robinson, H; Xin, Z; Lin, G; Ke, H Multiple conformations of phosphodiesterase-5: implications for enzyme function and drug development. J Biol Chem281:21469-79 (2006) [PubMed] Article
Wang, H; Liu, Y; Huai, Q; Cai, J; Zoraghi, R; Francis, SH; Corbin, JD; Robinson, H; Xin, Z; Lin, G; Ke, H Multiple conformations of phosphodiesterase-5: implications for enzyme function and drug development. J Biol Chem281:21469-79 (2006) [PubMed] Article