| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Inosine-5'-monophosphate dehydrogenase 1 |
|---|
| Ligand | BDBM19264 |
|---|
| Substrate/Competitor | BDBM19254 |
|---|
| Meas. Tech. | IMPDH Type 1 Enzyme Assay |
|---|
| pH | 8±n/a |
|---|
| Temperature | 298.15±n/a K |
|---|
| Ki | 33±n/a nM |
|---|
| Comments | K562 cell proliferation, IC50=7.7 uM. |
|---|
| Citation |  Chen, L; Gao, G; Felczak, K; Bonnac, L; Patterson, SE; Wilson, D; Bennett, EM; Jayaram, HN; Hedstrom, L; Pankiewicz, KW Probing Binding Requirements of Type I and Type II Isoforms of Inosine Monophosphate Dehydrogenase with Adenine-Modified Nicotinamide Adenine Dinucleotide Analogues. J Med Chem50:5743-5751 (2007) [PubMed] Article Chen, L; Gao, G; Felczak, K; Bonnac, L; Patterson, SE; Wilson, D; Bennett, EM; Jayaram, HN; Hedstrom, L; Pankiewicz, KW Probing Binding Requirements of Type I and Type II Isoforms of Inosine Monophosphate Dehydrogenase with Adenine-Modified Nicotinamide Adenine Dinucleotide Analogues. J Med Chem50:5743-5751 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Inhibition_Run data, Solution Info, Assay Method |
|---|
| |
| Inosine-5'-monophosphate dehydrogenase 1 |
|---|
| Name: | Inosine-5'-monophosphate dehydrogenase 1 |
|---|
| Synonyms: | IMDH1_HUMAN | IMP dehydrogenase 1 | IMPD 1 | IMPD1 | IMPDH-I | IMPDH1 | Inosine Monophosphate Dehydrogenase Type 1 (IMPDH1) | Inosine-5'-monophosphate dehydrogenase (IMPDH) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 55407.70 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Recombinant IMPDH1 expressed in E. coli. |
|---|
| Residue: | 514 |
|---|
| Sequence: | MADYLISGGTGYVPEDGLTAQQLFASADGLTYNDFLILPGFIDFIADEVDLTSALTRKIT
LKTPLISSPMDTVTEADMAIAMALMGGIGFIHHNCTPEFQANEVRKVKKFEQGFITDPVV
LSPSHTVGDVLEAKMRHGFSGIPITETGTMGSKLVGIVTSRDIDFLAEKDHTTLLSEVMT
PRIELVVAPAGVTLKEANEILQRSKKGKLPIVNDCDELVAIIARTDLKKNRDYPLASKDS
QKQLLCGAAVGTREDDKYRLDLLTQAGVDVIVLDSSQGNSVYQIAMVHYIKQKYPHLQVI
GGNVVTAAQAKNLIDAGVDGLRVGMGCGSICITQEVMACGRPQGTAVYKVAEYARRFGVP
IIADGGIQTVGHVVKALALGASTVMMGSLLAATTEAPGEYFFSDGVRLKKYRGMGSLDAM
EKSSSSQKRYFSEGDKVKIAQGVSGSIQDKGSIQKFVPYLIAGIQHGCQDIGARSLSVLR
SMMYSGELKFEKRTMSAQIEGGVHGLHSYEKRLY
|
|
|
|---|
| BDBM19264 |
|---|
| BDBM19254 |
|---|
| Name | BDBM19264 |
|---|
| Synonyms: | (4E)-6-(4-hydroxy-6-methoxy-7-methyl-3-oxo-1,3-dihydro-2-benzofuran-5-yl)-4-methylhex-4-enoic acid | (E)-6-(4-hydroxy-6-methoxy-7-methyl-3-oxo-1H-2-benzofuran-5-yl)-4-methylhex-4-enoic acid | CHEMBL866 | MPA (1) | Mycophenolic Acid (MPA) | Myfortic |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H20O6 |
|---|
| Mol. Mass. | 320.3371 |
|---|
| SMILES | COc1c(C)c2COC(=O)c2c(O)c1C\C=C(/C)CCC(O)=O |
|---|
| Structure | 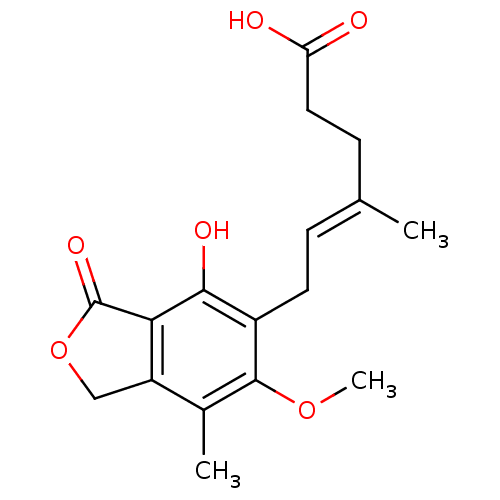
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Chen, L; Gao, G; Felczak, K; Bonnac, L; Patterson, SE; Wilson, D; Bennett, EM; Jayaram, HN; Hedstrom, L; Pankiewicz, KW Probing Binding Requirements of Type I and Type II Isoforms of Inosine Monophosphate Dehydrogenase with Adenine-Modified Nicotinamide Adenine Dinucleotide Analogues. J Med Chem50:5743-5751 (2007) [PubMed] Article
Chen, L; Gao, G; Felczak, K; Bonnac, L; Patterson, SE; Wilson, D; Bennett, EM; Jayaram, HN; Hedstrom, L; Pankiewicz, KW Probing Binding Requirements of Type I and Type II Isoforms of Inosine Monophosphate Dehydrogenase with Adenine-Modified Nicotinamide Adenine Dinucleotide Analogues. J Med Chem50:5743-5751 (2007) [PubMed] Article