| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Equilibrative nucleoside transporter 1 |
|---|
| Ligand | BDBM23638 |
|---|
| Substrate/Competitor | BDBM23618 |
|---|
| Meas. Tech. | Flow Cytometric Assay |
|---|
| pH | 7.4±n/a |
|---|
| Temperature | 295.15±n/a K |
|---|
| Comments | 16.2 +/- 1.9 % Inhibition @ 10 uM. |
|---|
| Citation |  Lin, W; Buolamwini, JK Synthesis, flow cytometric evaluation, and identification of highly potent dipyridamole analogues as equilibrative nucleoside transporter 1 inhibitors. J Med Chem50:3906-20 (2007) [PubMed] Article Lin, W; Buolamwini, JK Synthesis, flow cytometric evaluation, and identification of highly potent dipyridamole analogues as equilibrative nucleoside transporter 1 inhibitors. J Med Chem50:3906-20 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Equilibrative nucleoside transporter 1 |
|---|
| Name: | Equilibrative nucleoside transporter 1 |
|---|
| Synonyms: | ENT1 | Equilibrative NBMPR-sensitive nucleoside transporter | Equilibrative Nucleoside Transporter 1 (ENT1) | Equilibrative nitrobenzylmercaptopurine riboside-sensitive nucleoside transporter | Equilibrative nucleoside transporter 1 | Nucleoside transporter, es-type | S29A1_HUMAN | SLC29A1 | Solute carrier family 29 member 1 |
|---|
| Type: | Multi-pass membrane protein |
|---|
| Mol. Mass.: | 50225.92 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 456 |
|---|
| Sequence: | MTTSHQPQDRYKAVWLIFFMLGLGTLLPWNFFMTATQYFTNRLDMSQNVSLVTAELSKDA
QASAAPAAPLPERNSLSAIFNNVMTLCAMLPLLLFTYLNSFLHQRIPQSVRILGSLVAIL
LVFLITAILVKVQLDALPFFVITMIKIVLINSFGAILQGSLFGLAGLLPASYTAPIMSGQ
GLAGFFASVAMICAIASGSELSESAFGYFITACAVIILTIICYLGLPRLEFYRYYQQLKL
EGPGEQETKLDLISKGEEPRAGKEESGVSVSNSQPTNESHSIKAILKNISVLAFSVCFIF
TITIGMFPAVTVEVKSSIAGSSTWERYFIPVSCFLTFNIFDWLGRSLTAVFMWPGKDSRW
LPSLVLARLVFVPLLLLCNIKPRRYLTVVFEHDAWFIFFMAAFAFSNGYLASLCMCFGPK
KVKPAEAETAGAIMAFFLCLGLALGAVFSFLFRAIV
|
|
|
|---|
| BDBM23638 |
|---|
| BDBM23618 |
|---|
| Name | BDBM23638 |
|---|
| Synonyms: | 2-{[4,8-bis({4-azatricyclo[4.3.1.1^{3,8}]undecan-4-yl})-6-[(2-hydroxyethyl)amino]pyrimido[5,4-d][1,3]diazin-2-yl]amino}ethan-1-ol | Dipyridamole Analogue, 20 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C30H44N8O2 |
|---|
| Mol. Mass. | 548.7228 |
|---|
| SMILES | OCCNc1nc(N2CC3CC4CC(C3)CC2C4)c2nc(NCCO)nc(N3CC4CC5CC(C4)CC3C5)c2n1 |TLB:8:9:17.16.15:12,14:13:17:10.9.8.7,14:9:17:15.12.13,28:29:37.36.35:32,34:33:37:30.29.28.27,34:29:37:35.32.33,THB:26:27:37:35.32.33,6:7:17:15.12.13,7:16:12:10.9.14,27:36:32:30.29.34| |
|---|
| Structure | 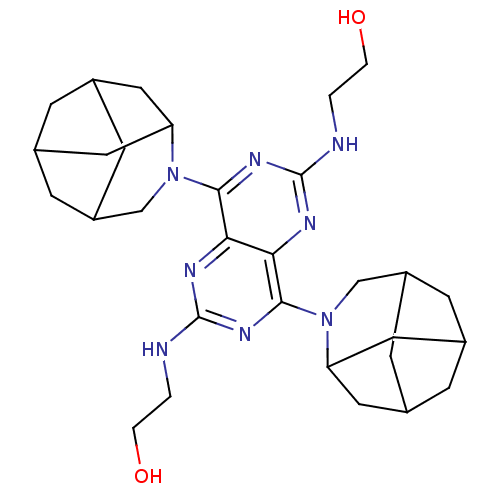
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lin, W; Buolamwini, JK Synthesis, flow cytometric evaluation, and identification of highly potent dipyridamole analogues as equilibrative nucleoside transporter 1 inhibitors. J Med Chem50:3906-20 (2007) [PubMed] Article
Lin, W; Buolamwini, JK Synthesis, flow cytometric evaluation, and identification of highly potent dipyridamole analogues as equilibrative nucleoside transporter 1 inhibitors. J Med Chem50:3906-20 (2007) [PubMed] Article