

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Epoxide hydrolase B | ||
| Ligand | BDBM25742 | ||
| Substrate/Competitor | BDBM25726 | ||
| Meas. Tech. | Mtb EHB Inhibition Assay | ||
| pH | 7±n/a | ||
| Temperature | 303.15±n/a K | ||
| IC50 | 2060±n/a nM | ||
| Citation |  Biswal, BK; Morisseau, C; Garen, G; Cherney, MM; Garen, C; Niu, C; Hammock, BD; James, MN The molecular structure of epoxide hydrolase B from Mycobacterium tuberculosis and its complex with a urea-based inhibitor. J Mol Biol381:897-912 (2008) [PubMed] Article Biswal, BK; Morisseau, C; Garen, G; Cherney, MM; Garen, C; Niu, C; Hammock, BD; James, MN The molecular structure of epoxide hydrolase B from Mycobacterium tuberculosis and its complex with a urea-based inhibitor. J Mol Biol381:897-912 (2008) [PubMed] Article | ||
| More Info.: | Get all data from this article, Inhibition_Run data, Solution Info, Assay Method | ||
| Epoxide hydrolase B | |||
| Name: | Epoxide hydrolase B | ||
| Synonyms: | EPHB_MYCTO | Epoxide Hydrolase B (EHB) | Mtb EHB | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 39285.78 | ||
| Organism: | Mycobacterium tuberculosis | ||
| Description: | The His6-tagged Mtb EHB was expressed in E. coli and purified. | ||
| Residue: | 356 | ||
| Sequence: |
| ||
| BDBM25742 | |||
| BDBM25726 | |||
| Name | BDBM25742 | ||
| Synonyms: | 1-[4-(4-fluorophenoxy)cyclohexyl]-3-[4-(trifluoromethoxy)phenyl]urea | Urea-based compound, 23 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C20H20F4N2O3 | ||
| Mol. Mass. | 412.378 | ||
| SMILES | Fc1ccc(OC2CCC(CC2)NC(=O)Nc2ccc(OC(F)(F)F)cc2)cc1 |(13.37,-2.95,;12.06,-2.13,;10.72,-2.9,;9.39,-2.13,;9.39,-.59,;7.9,-.2,;7.9,1.34,;6.86,2.47,;5.01,1.58,;3.75,1.86,;4.78,.7,;6.61,1.63,;2.42,1.09,;1.08,1.86,;1.08,3.4,;-.25,1.08,;-1.58,1.85,;-2.92,1.08,;-4.25,1.85,;-4.25,3.4,;-5.58,4.17,;-6.92,3.39,;-8.25,4.16,;-8.25,2.62,;-6.92,1.85,;-2.92,4.17,;-1.58,3.4,;10.72,.18,;12.15,-.58,)| | ||
| Structure | 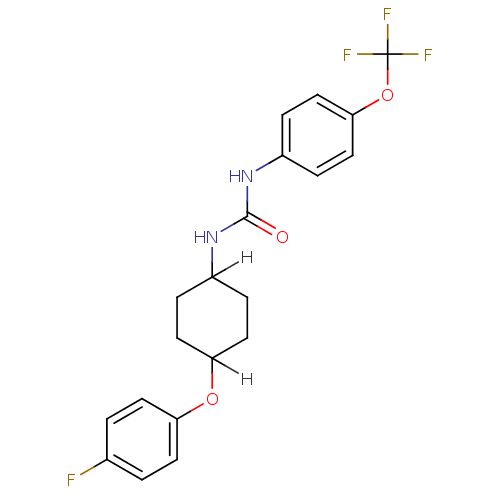
| ||