| Reaction Details |
|---|
 | Report a problem with these data |
| Target | G2/mitotic-specific cyclin-B1 |
|---|
| Ligand | BDBM43728 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Counterscreen assay for inhibitors of Wee1 degradation: dose response cell-based assay to identify inhibitors of cyclin B degradation |
|---|
| EC50 | >49750±n/a nM |
|---|
| Citation |  PubChem, PC Counterscreen assay for inhibitors of Wee1 degradation: dose response cell-based assay to identify inhibitors of cyclin B degradation PubChem Bioassay(2008)[AID] PubChem, PC Counterscreen assay for inhibitors of Wee1 degradation: dose response cell-based assay to identify inhibitors of cyclin B degradation PubChem Bioassay(2008)[AID] |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| G2/mitotic-specific cyclin-B1 |
|---|
| Name: | G2/mitotic-specific cyclin-B1 |
|---|
| Synonyms: | CCNB | CCNB1 | CCNB1_HUMAN |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 48340.95 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P14635 |
|---|
| Residue: | 433 |
|---|
| Sequence: | MALRVTRNSKINAENKAKINMAGAKRVPTAPAATSKPGLRPRTALGDIGNKVSEQLQAKM
PMKKEAKPSATGKVIDKKLPKPLEKVPMLVPVPVSEPVPEPEPEPEPEPVKEEKLSPEPI
LVDTASPSPMETSGCAPAEEDLCQAFSDVILAVNDVDAEDGADPNLCSEYVKDIYAYLRQ
LEEEQAVRPKYLLGREVTGNMRAILIDWLVQVQMKFRLLQETMYMTVSIIDRFMQNNCVP
KKMLQLVGVTAMFIASKYEEMYPPEIGDFAFVTDNTYTKHQIRQMEMKILRALNFGLGRP
LPLHFLRRASKIGEVDVEQHTLAKYLMELTMLDYDMVHFPPSQIAAGAFCLALKILDNGE
WTPTLQHYLSYTEESLLPVMQHLAKNVVMVNQGLTKHMTVKNKYATSKHAKISTLPQLNS
ALVQDLAKAVAKV
|
|
|
|---|
| BDBM43728 |
|---|
| n/a |
|---|
| Name | BDBM43728 |
|---|
| Synonyms: | (E)-N-(5-chloranylpyridin-2-yl)-3-phenyl-prop-2-enamide | (E)-N-(5-chloro-2-pyridinyl)-3-phenyl-2-propenamide | (E)-N-(5-chloro-2-pyridyl)-3-phenyl-acrylamide | (E)-N-(5-chloropyridin-2-yl)-3-phenylprop-2-enamide | MLS000684215 | N-(5-chloro-2-pyridinyl)-3-phenylacrylamide | SMR000268017 | cid_691131 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H11ClN2O |
|---|
| Mol. Mass. | 258.703 |
|---|
| SMILES | Clc1ccc(NC(=O)\C=C\c2ccccc2)nc1 |
|---|
| Structure | 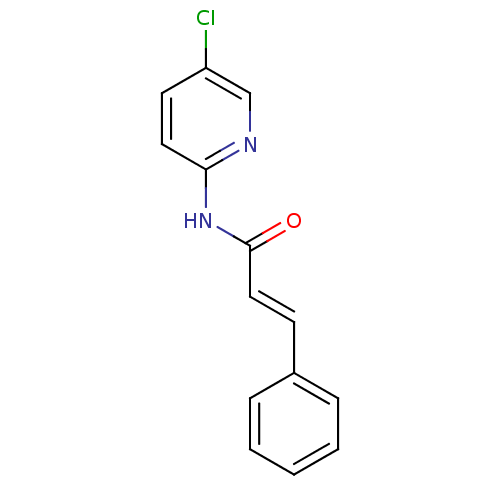
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Counterscreen assay for inhibitors of Wee1 degradation: dose response cell-based assay to identify inhibitors of cyclin B degradation PubChem Bioassay(2008)[AID]
PubChem, PC Counterscreen assay for inhibitors of Wee1 degradation: dose response cell-based assay to identify inhibitors of cyclin B degradation PubChem Bioassay(2008)[AID]