| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Induced myeloid leukemia cell differentiation protein Mcl-1 |
|---|
| Ligand | BDBM35379 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Dose Response Confirmation for Mcl-1/Noxa Interaction Inhibitors |
|---|
| IC50 | 2591.202711±n/a nM |
|---|
| Citation |  PubChem, PC Dose Response Confirmation for Mcl-1/Noxa Interaction Inhibitors PubChem Bioassay(2008)[AID] PubChem, PC Dose Response Confirmation for Mcl-1/Noxa Interaction Inhibitors PubChem Bioassay(2008)[AID] |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Induced myeloid leukemia cell differentiation protein Mcl-1 |
|---|
| Name: | Induced myeloid leukemia cell differentiation protein Mcl-1 |
|---|
| Synonyms: | BCL2L3 | Bcl-2-like protein 3 | Bcl-2-like protein 3 (Mcl-1) | Bcl-2-related protein EAT/mcl1 | Bcl2-L-3 | Induced myeloid leukemia cell differentiation protein (Mcl-1) | MCL1 | MCL1_HUMAN | Mcl-1 | Myeloid Cell factor-1 (Mcl-1) | Myeloid cell leukemia sequence 1 (BCL2-related) | mcl1/EAT |
|---|
| Type: | Membrane; Single-pass membrane protein |
|---|
| Mol. Mass.: | 37332.87 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q07820 |
|---|
| Residue: | 350 |
|---|
| Sequence: | MFGLKRNAVIGLNLYCGGAGLGAGSGGATRPGGRLLATEKEASARREIGGGEAGAVIGGS
AGASPPSTLTPDSRRVARPPPIGAEVPDVTATPARLLFFAPTRRAAPLEEMEAPAADAIM
SPEEELDGYEPEPLGKRPAVLPLLELVGESGNNTSTDGSLPSTPPPAEEEEDELYRQSLE
IISRYLREQATGAKDTKPMGRSGATSRKALETLRRVGDGVQRNHETAFQGMLRKLDIKNE
DDVKSLSRVMIHVFSDGVTNWGRIVTLISFGAFVAKHLKTINQESCIEPLAESITDVLVR
TKRDWLVKQRGWDGFVEFFHVEDLEGGIRNVLLAFAGVAGVGAGLAYLIR
|
|
|
|---|
| BDBM35379 |
|---|
| n/a |
|---|
| Name | BDBM35379 |
|---|
| Synonyms: | 2-(2-furanyl)-4-quinolinecarboxylic acid [2-[4-amino-1-methyl-3-(2-methylpropyl)-2,6-dioxo-5-pyrimidinyl]-2-oxoethyl] ester | 2-(2-furyl)cinchoninic acid [2-(6-amino-1-isobutyl-2,4-diketo-3-methyl-pyrimidin-5-yl)-2-keto-ethyl] ester | MLS000058168 | SMR000063262 | [2-[4-amino-1-methyl-3-(2-methylpropyl)-2,6-dioxopyrimidin-5-yl]-2-oxoethyl] 2-(furan-2-yl)quinoline-4-carboxylate | [2-[4-azanyl-1-methyl-3-(2-methylpropyl)-2,6-bis(oxidanylidene)pyrimidin-5-yl]-2-oxidanylidene-ethyl] 2-(furan-2-yl)quinoline-4-carboxylate | cid_2094474 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C25H24N4O6 |
|---|
| Mol. Mass. | 476.4813 |
|---|
| SMILES | CC(C)Cn1c(N)c(C(=O)COC(=O)c2cc(nc3ccccc23)-c2ccco2)c(=O)n(C)c1=O |
|---|
| Structure | 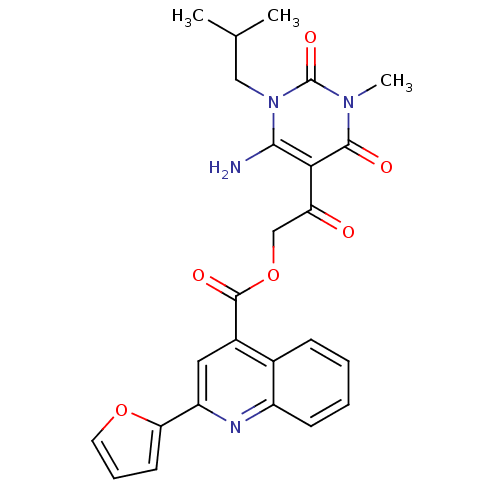
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Dose Response Confirmation for Mcl-1/Noxa Interaction Inhibitors PubChem Bioassay(2008)[AID]
PubChem, PC Dose Response Confirmation for Mcl-1/Noxa Interaction Inhibitors PubChem Bioassay(2008)[AID]