| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Alkaline phosphatase, tissue-nonspecific isozyme |
|---|
| Ligand | BDBM59258 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | uHTS identification of compounds activating TNAP in the absence of phosphate acceptor performed in luminescent assay |
|---|
| EC50 | 100000±n/a nM |
|---|
| Citation |  PubChem, PC uHTS identification of compounds activating TNAP in the absence of phosphate acceptor performed in luminescent assay PubChem Bioassay(2009)[AID] PubChem, PC uHTS identification of compounds activating TNAP in the absence of phosphate acceptor performed in luminescent assay PubChem Bioassay(2009)[AID] |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Alkaline phosphatase, tissue-nonspecific isozyme |
|---|
| Name: | Alkaline phosphatase, tissue-nonspecific isozyme |
|---|
| Synonyms: | ALPL | PPBT_HUMAN | tissue-nonspecific alkaline phosphatase precursor |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 57306.17 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_1448917 |
|---|
| Residue: | 524 |
|---|
| Sequence: | MISPFLVLAIGTCLTNSLVPEKEKDPKYWRDQAQETLKYALELQKLNTNVAKNVIMFLGD
GMGVSTVTAARILKGQLHHNPGEETRLEMDKFPFVALSKTYNTNAQVPDSAGTATAYLCG
VKANEGTVGVSAATERSRCNTTQGNEVTSILRWAKDAGKSVGIVTTTRVNHATPSAAYAH
SADRDWYSDNEMPPEALSQGCKDIAYQLMHNIRDIDVIMGGGRKYMYPKNKTDVEYESDE
KARGTRLDGLDLVDTWKSFKPRYKHSHFIWNRTELLTLDPHNVDYLLGLFEPGDMQYELN
RNNVTDPSLSEMVVVAIQILRKNPKGFFLLVEGGRIDHGHHEGKAKQALHEAVEMDRAIG
QAGSLTSSEDTLTVVTADHSHVFTFGGYTPRGNSIFGLAPMLSDTDKKPFTAILYGNGPG
YKVVGGERENVSMVDYAHNNYQAQSAVPLRHETHGGEDVAVFSKGPMAHLLHGVHEQNYV
PHVMAYAACIGANLGHCAPASSAGSLAAGPLLLALALYPLSVLF
|
|
|
|---|
| BDBM59258 |
|---|
| n/a |
|---|
| Name | BDBM59258 |
|---|
| Synonyms: | (5E)-5-(6-ketocyclohexa-2,4-dien-1-ylidene)-N-p-anisyl-3-pyrazoline-3-carboxamide | (5E)-N-[(4-methoxyphenyl)methyl]-5-(6-oxidanylidenecyclohexa-2,4-dien-1-ylidene)-1,2-dihydropyrazole-3-carboxamide | (5E)-N-[(4-methoxyphenyl)methyl]-5-(6-oxo-1-cyclohexa-2,4-dienylidene)-1,2-dihydropyrazole-3-carboxamide | (5E)-N-[(4-methoxyphenyl)methyl]-5-(6-oxocyclohexa-2,4-dien-1-ylidene)-1,2-dihydropyrazole-3-carboxamide | MLS000084094 | SMR000047293 | cid_5389985 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H17N3O3 |
|---|
| Mol. Mass. | 323.3459 |
|---|
| SMILES | COc1ccc(CNC(=O)c2cc(n[nH]2)-c2ccccc2O)cc1 |
|---|
| Structure | 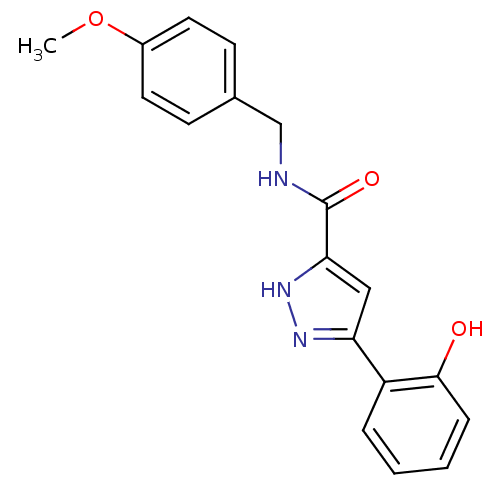
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC uHTS identification of compounds activating TNAP in the absence of phosphate acceptor performed in luminescent assay PubChem Bioassay(2009)[AID]
PubChem, PC uHTS identification of compounds activating TNAP in the absence of phosphate acceptor performed in luminescent assay PubChem Bioassay(2009)[AID]